Deploy Language Models with OpenVINO™ Model Server
Authors: Dariusz Trawinski, Damian Kalinowski
Overview
If you are writing an AI application that handles text in Natural Language Processing (NLP) models, you will be pleased to hear that OpenVINO Model Server now supports sending and receiving text in string format.
Now you can combine optimized inference execution with a simple method for sending text data to the model server and reading text responses.
Introduction
Deep Learning models do not deal with text content directly. Instead, they require a numerical representation of text to process it.
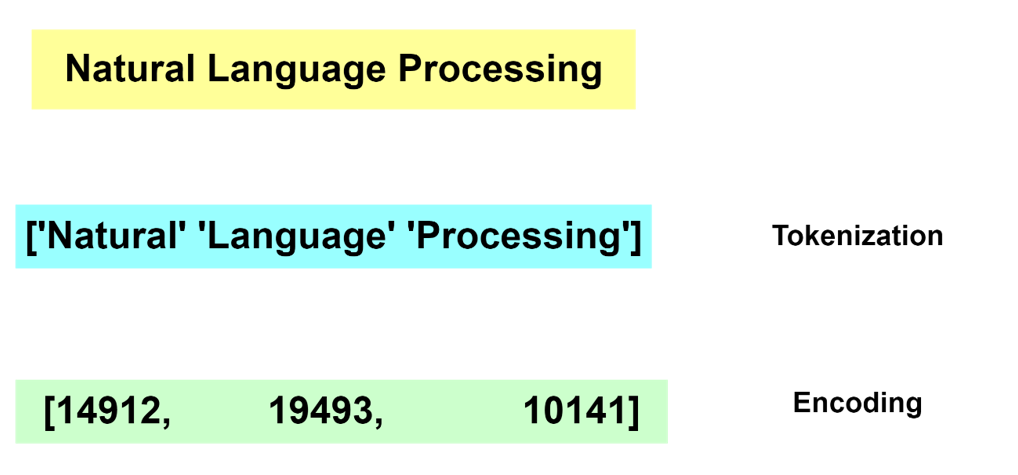
The conversion from human readable text to a machine-readable format is done via a process of tokenization and encoding. Without going into the specifics of tokenization and encoding, these operations are not trivial. Many algorithms exist for these tasks and most often the operation is run by dedicated software libraries.
Generally, during the inference operation, a client application must reproduce the same method for text tokenization and encoding, similar to what is used during the model training phase.
For reference, below are two examples showing how this can be implemented on the application side as pre- and post-processing steps:
In TensorFlow it’s also possible to embed the tokenization operation inside the model by adding a dedicated neuron model layer SentencePieceTokenizer.
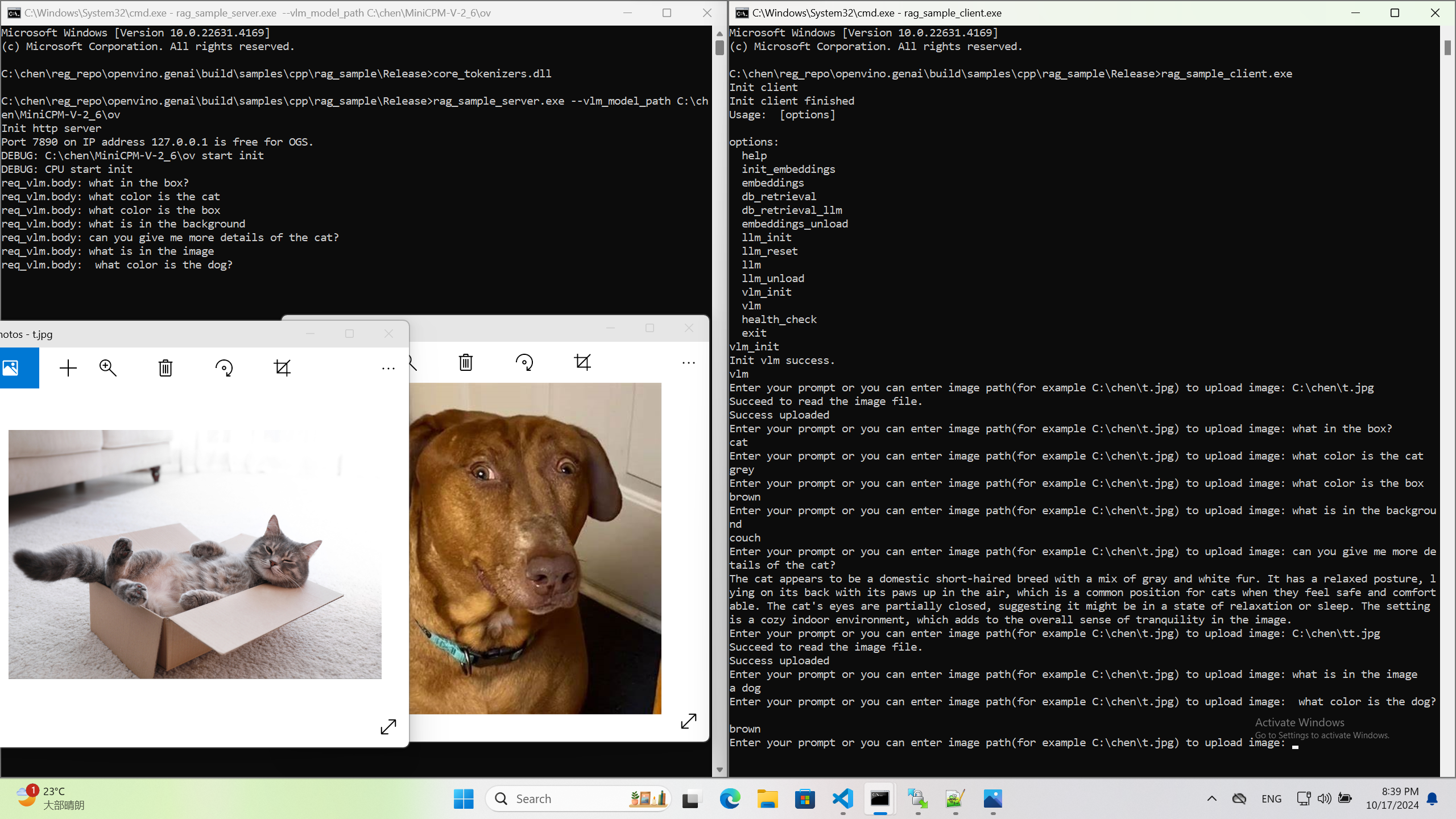
Tokenization and Encoding with OpenVINO Model Server
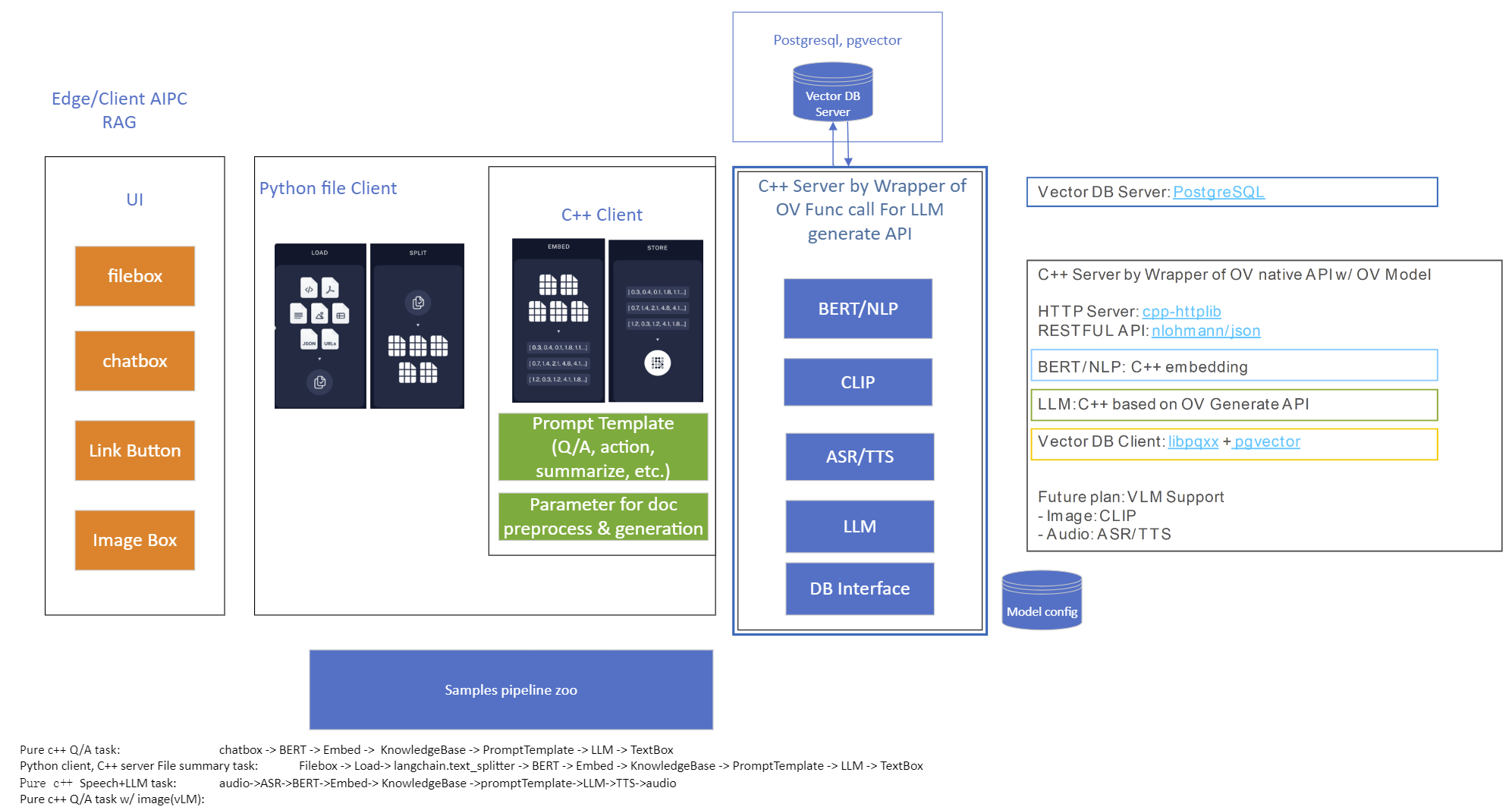
Starting with the 2023.0 release, OpenVINO Model Server can greatly simplify writing applications that leverage LLM and NLP models. We addressed both using models that require tokens and models with an embedded tokenization layer. Both use cases are demonstrated below with a simple client application that sends and receives text in a string format. The complexity of text conversion is fully delegated to the remote serving endpoint.
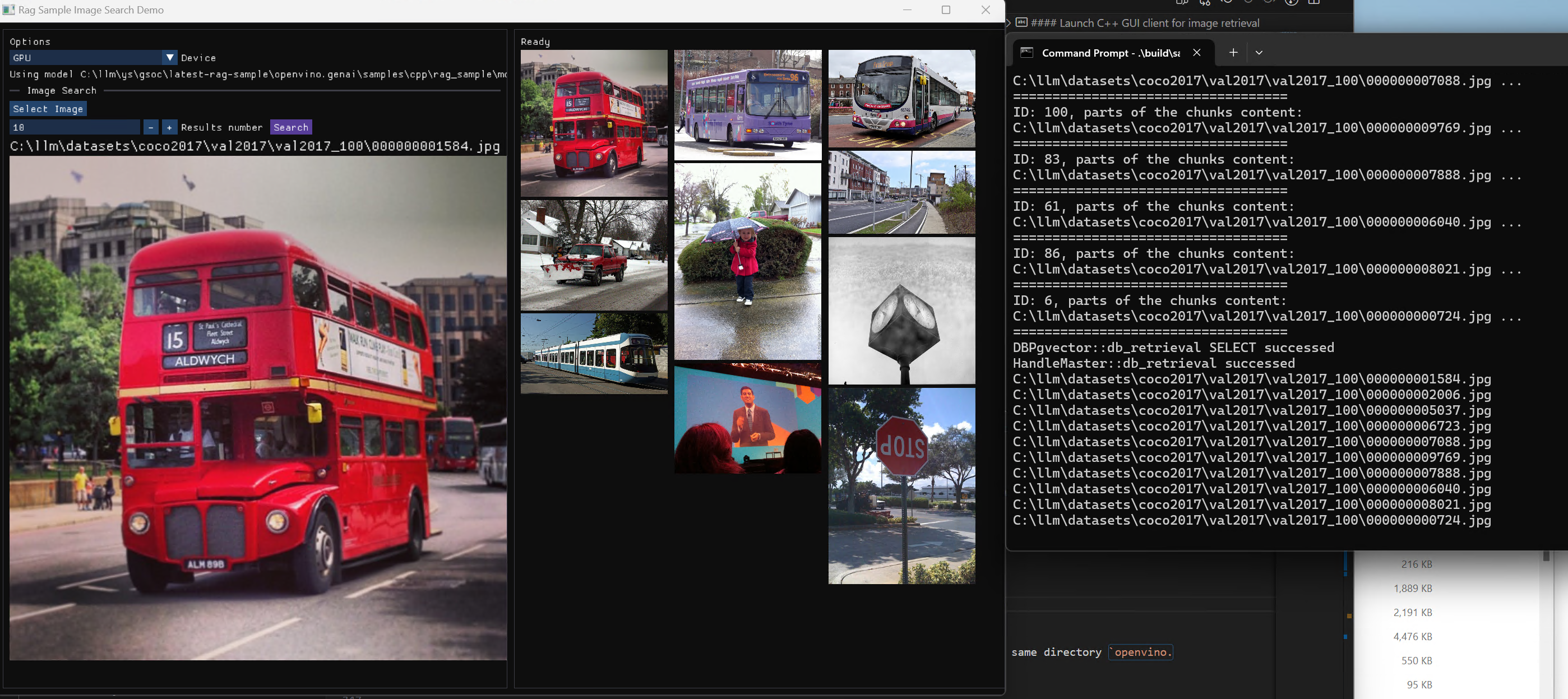
GPT-J Pipeline
In this demo we deploy the tokenizer as a custom node in OpenVINO Model Server. As a result, we get a pipeline with seed strings as input and generated texts as the output.
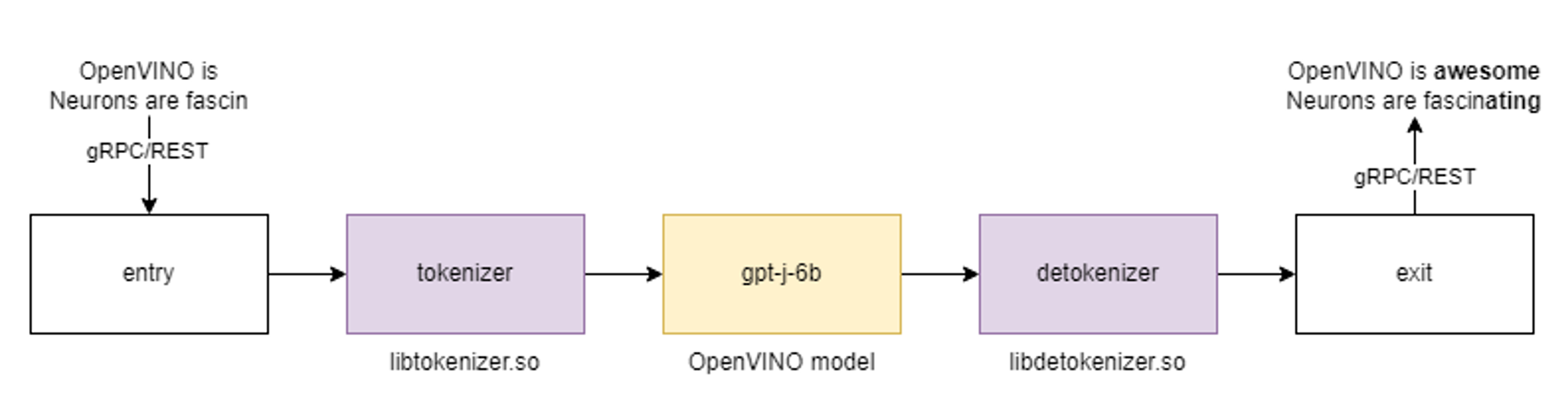
All steps to reproduce the demo above are documented here: https://docs.openvino.ai/2023.0/ovms_demo_gptj_causal_lm.html
Text generation can be executed iteratively in a loop. An example of the client application generating text output is shown below.

Multilingual Universal Sentence Encoder (MUSE)
The next demonstration includes serving the MUSE model from TensorFlow Hub. The demo shows how OpenVINO Model Server can be used to serve the MUSE model and with 2x better performance without any changes on the client side.
The calls to the model server are simple using a REST API. Below is an example of a call with a batch size 3.
A similar call can be made over gRPC interface using the ovmsclient library which is compatible with the TensorFlow Serving (TFS) API.
In addition to the TFS API, it is also possible to run inference calls using the KServe v2 API. Check the code snippets for more details.
Conclusion
OpenVINO Model Server can simplify writing AI applications that handle text. It can execute a complete text analysis pipeline with just few[TA4] lines of code on the client side without compromising performance by using a tokenizer in C++ and high performance OpenVINO backend to run the AI models. Together with widely used, standard APIs, OpenVINO Model Server is a great solution for deploying effective and efficient AI applications.
References
https://towardsdatascience.com/tokenization-algorithms-explained-e25d5f4322ac
https://www.tensorflow.org/api_docs/python/tfm/nlp/layers/SentencepieceTokenizer
https://docs.openvino.ai/2023.0/ovms_what_is_openvino_model_server.html
https://docs.openvino.ai/2023.0/openvino_docs_performance_benchmarks.html

.png)