An end-to-end workflow with training on Habana® Gaudi® Processor and post-training quantization and Inference using OpenVINO™ toolkit
Authors: Sachin Rastogi, Maajid N Khan, Akhila Vidiyala
Background:
Brain tumors are abnormal growths of braincells and can be benign (non-cancerous) or malignant (cancerous). Accurate diagnosis and treatment of brain tumors are critical for the patient's prognosis, and one important step in this process is the segmentation of the tumor in medical images. This involves identifying the boundaries of the tumor and separating it from the surrounding healthy brain tissue.
MRI is a non-invasive imaging technique that uses a strong magnetic field and radio waves to produce detailed images of the brain. MRI scans can provide high-resolution images of the brain, including the location and size of tumors. Traditionally, trained professionals, such as radiologists or medical image analysts, perform manual segmentation of brain tumors. However, this process is time-consuming and subject to human error, leading to the development of automated methods using machine learning.
Introduction:
As demand for deep learning applications grows for medical imaging, so does the need for cost-effective cloud solutions that can efficiently train Deep Learning models. With the Amazon EC2 DL1 instances powered by Gaudi® accelerators from Habana® Labs (An Intel® company), you can train deep learning models for medical image segmentation at a reduced cost of up to 40% than the current generation GPU-based EC2 instances.
Medical Imaging AI solutions often need to be deployed on various hardware platforms, including both new and older systems. The usage of Intel® Distribution of OpenVINO™ toolkit makes it easier to deploy these solutions on systems with Intel® Architecture.
This reference implementation demonstrates how this toolkit can be used to detect and distinguish between healthy and cancerous tissue in MRI scans. It can be used on a range of Intel® Architecture platforms, including CPUs, integrated GPUs, and VPUs, with no need to modify the code when switching between platforms. This allows developers to choose the hardware that meets their needs in terms of performance, cost, and power consumption.
The Challenge:
Identify and separate cancerous tumors from healthy tissue in an MRI scan of the brain with the best price performance.
The Solution:
One approach to brain tumor segmentation using machine learning is to use supervised learning, where the algorithm is trained on a dataset of labelled brain images, with the tumor regions already identified by experts. The algorithm can then learn to identify these tumor regions in new images.
Convolutional neural networks (CNNs) are a type of machine learning model that has been successful in image classification and segmentation tasks and are often used for brain tumor segmentation. In a CNN, the input image is passed through multiple layers of filters that learn to recognize specific features in the image. The output of the CNN is a segmented image, with each pixel classified as either part of the tumor or healthy tissue.
Another approach to brain tumor segmentation is to use unsupervised learning, where the algorithm is not given any labelled examples and must learn to identify patterns in the data on its own. One unsupervised method for brain tumor segmentation is to use clustering algorithms, which can group similar pixels together and identify the tumor region as a separate cluster. However, unsupervised learning is not commonly used for brain tumor segmentation due to the complexity and variability of the data.
Regardless of the approach used, the performance of brain tumor segmentation algorithms can be evaluated using metrics such as dice coefficient, Jaccard index, and sensitivity.
Our medical imaging AI solution is designed to be used widely and in a cost-effective manner. Our approach ensures that the accuracy of the model is not compromised while still being affordable. We have used a U-Net 2D model that can be trained using the Habana® Gaudi® platform and the Medical Decathlon dataset (BraTS 2017 Brain Tumor Dataset) to achieve the best possible accuracy for image segmentation. The model can then be used for inferencing with the OpenVINO™ on Intel® Architecture.
This reference implementation provides an AWS*cloud-based generic AI workflow, which showcases U-Net-2D model-based image segmentation with the medical decathlon dataset. The reference implementation is available for use by Docker containers and Helm chart.

Training:
Primarily, we are leveraging AWS* EC2 DL1workflows to train U-Net 2D models for the end-to-end pipeline. We are consistently seeing cost savings compared to existing GPU-based instances across model types, enabling us to achieve much better Time-to-Market for existing models or training much larger and more complex models.
AWS*DL1 instances with Gaudi® accelerators offer the best price-performance savings compared to other GPU offerings in the market. The models were trained using the Pytorch framework.
The reference training code with detailed instructions is available here.
Inference and Optimization:
Intel® OpenVINO™ is an inference solution that optimizes and accelerates the computation of AI workloads on Intel® hardware. The trained Pytorch models were converted to ONNX (Open Neural Network Exchange) model representation format and then further optimized to the OpenVINO™ format or Intermediate representation (IR) of OpenVINO™ using the Model Optimizer tool from OpenVINO™.
TheFP32-optimized IR models outperformed using OpenVINO™ runtime in terms of throughput compared to other Deep Learning framework runtimes on the same Intel® hardware.
Asa next step, the FP32 IR model was further optimized and converted to lower8-bit precision with post-training quantization using the default quantization algorithm from the Post Training Optimization Tool (POT) from the OpenVINO™ toolkit. This inherently leads to a jump in the model’s performance, in terms of its processing speed and throughput, for you get a higher FPS when dealing with video streams with very negligible loss in accuracy.
TheINT8 IR models performed extremely well for inference on Intel® CPU(Central Processing Unit) 3rd Generation Intel® Xeon.
The reference inference code with detailed instructions is available here.
GitHub: https://github.com/intel/cv-training-and-inference-openvino/tree/main/gaudi-segmentation-unet-ptq
Developer Catalog: https://www.intel.com/content/www/us/en/developer/articles/reference-implementation/gaudi-processor-training-and-inference-openvino.html
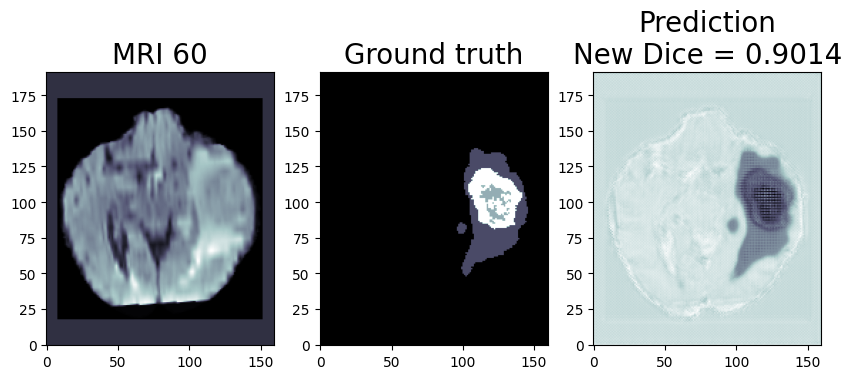
Inference Result:
We are using OpenVINO™ Model Optimizer(MO) to convert the trained ONNX FP32 model to FP32 OpenVINO™ or Intermediate Representation(IR) format. The FP32prediction shown here is from a test image from the training dataset which was never used for training. The prediction is from a trained model which was trained for 8 epochs with 8 HPU multi-card training on an AWS* EC2 DL1 Instance with 400/484 images from the training folder.
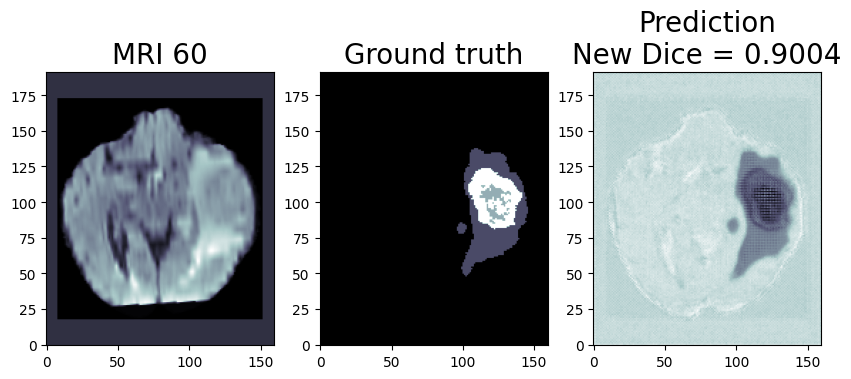
Quantization (Recommended to use if you need the better performance of the model)
Quantization is the process of converting a deep learning model’s weight to a lower precision requiring less computation. This inherently leads to an increase in model performance, in terms of its processing speed and throughput, you will see a higher throughput(FPS) when dealing with video streams. We are using OpenVINO™ POT for the Default Quantization Algorithm to quantize the FP32 OpenVINO™ format model into the INT8 OpenVINO™ format model.
The INT8 prediction shown here is from a testimage from a training dataset that was never used for training. The predictionis from a quantized model which we quantized using POT with a calibrationdataset of 300 samples.
This application is available on the Intel® Developer Catalog for the developers to use as it is or use as a base code to bootstrap their customized solution. Intel® Developer Catalog offers reference implementations and software packages to build modular applications using containerized building blocks. Using the containerized building blocks the developers can rapidly develop deployable solutions.
Conclusion:
In conclusion, brain tumor segmentation using machine learning can help improve the accuracy and efficiency of the diagnosis and treatment of brain tumors.
There are several challenges and limitations to using machine learning for brain tumor segmentation. One of the main challenges is the limited availability of annotated data, as it is time consuming and expensive to annotate large datasets of medical images. In addition, there is a high degree of variability and complexity in the data, as brain tumors can have different shapes, sizes, and intensity patterns on MRI scans. This can make it difficult for the machine learning algorithm to generalize and accurately classify tumors in new data.
Another challenge is the potential for bias in the training data, as the dataset may not be representative of the entire population. This can lead to inaccurate or biased results if the algorithm is not properly trained or validated.
While there are still challenges to be overcome, the use of machine learning in medical image analysis shows great promise for improving patient care.
Notices & Disclaimers:
Intel technologies may require enabled hardware, software or service activation.
No product or component can be absolutely secure.
Your costs and results may vary.
© Intel Corporation. Intel, the Intel logo, and other Intel marks are trademarks of Intel Corporation or its subsidiaries. Other names and brands may be claimed as the property of others.

.png)