OpenVINO Blog
Ollama Integrated with OpenVINO, Accelerating DeepSeek Inference
Authors: Hongbo Zhao, Fiona Zhao, Tong Qiu
Why Choose the Ollama + OpenVINO Combination?
Dual-Engine Driven Technical Advantages
The integration of Ollama and OpenVINO delivers a powerful dual-engine solution for the management and inference of large language models (LLMs). Ollama offers a streamlined model management toolchain, while OpenVINO provides efficient acceleration capabilities for model inference across Intel hardware (CPU/GPU/NPU). This combination not only simplifies the deployment and invocation of models but also significantly enhances inference performance, making it particularly suitable for scenarios demanding high performance and ease of use.
You can find more information on github repository:
https://github.com/openvinotoolkit/openvino_contrib/tree/master/modules/ollama_openvino
Core Value of Ollama
1. Streamlined LLM Management Toolchain: Ollama provides a user-friendly command-line interface, enabling users to effortlessly download, manage, and run various LLM models.
2. One-Click Model Deployment: With simple commands, users can quickly deploy and invoke models without complex configurations.
3. Unified API Interface: Ollama offers a unified API interface, making it easy for developersto integrate into various applications.
4. Active Open-Source Community: Ollama boasts a vibrant open-source community, providing users with abundant resources and support.
Limitations of Ollama
Currently, Ollama only supports llama.cpp as itsbackend, which presents some inconveniences:
1. Limited Hardware Compatibility: llama.cpp is primarily optimized for CPUs and NVIDIA GPUs, and cannot fully leverage the acceleration capabilities of Intel GPUs or NPUs, resulting in suboptimal performance in high-performance computing scenarios.
2. Performance Bottlenecks: For large-scale models or high-concurrency scenarios, the performance of llama.cpp may fall short, especially when handling complex tasks, leading to slower inference speeds.
Breakthrough Capabilities of OpenVINO
1. Deep Optimization for Intel Hardware (CPU/iGPU/Arc dGPU/NPU): OpenVINO is deeply optimized for Intel hardware, fully leveraging the performance potential of CPUs, iGPUs, dGPUs, and NPUs.
2. Cross-Platform Heterogeneous Computing Support: OpenVINO supports cross-platform heterogeneous computing, enabling efficient model inference across different hardware platforms.
3. Model Quantization and Compression Toolchain: OpenVINO provides a comprehensive toolchain for model quantization and compression, significantly reducing model size and improving inference speed.
4. Significant Inference Performance Improvement: Through OpenVINO's optimizations, model inference performance can be significantly enhanced, especially for large-scale models and high-concurrency scenarios.
5. Extensibility and Flexibility Support: OpenVINO GenAI offers robust extensibility and flexibility for Ollama-OV, supporting pipeline optimization techniques such as speculative decoding, prompt-lookup decoding, pipeline parallelization, and continuous batching, laying a solid foundation for future pipeline serving optimizations.
Developer Benefits of Integration
1. Simplified Development Experience: Retains Ollama's CLI interaction features, allowing developers to continue using familiar command-line tools for model management and invocation.
2. Performance Leap: Achieves hardware-level acceleration through OpenVINO, significantly boosting model inference performance, especially for large-scale models and high-concurrency scenarios.
3. Multi-Hardware Adaptation and Ecosystem Expansion: OpenVINO's support enables Ollama to adapt to multiple hardware platforms, expanding its application ecosystem and providing developers with more choices and flexibility.
Three Steps to Enable Acceleration
1. Download Precompiled Executables
please refer to : https://github.com/zhaohb/ollama_ov/tree/main?tab=readme-ov-file#google-driver
2.Configure OpenVINO GenAI Environment
For Windows systems, first extract the downloaded OpenVINO GenAI package to the directory openvino_genai_windows_2025.2.0.0.dev20250320_x86_64, then execute the following commands:
cd openvino_genai_windows_2025.2.0.0.dev20250320_x86_64
setupvars.bat
3. Set Up cgocheck
Windows:
set GODEBUG=cgocheck=0
Linux:
export GODEBUG=cgocheck=0
At this point, the executable files have been downloaded, and the OpenVINO GenAI, OpenVINO, and CGO environments have been successfully configured.
Custom Model Deployment Guide
Since the Ollama Model Library does not support uploading non-GGUF format IR models, we will create an OCI image locally using OpenVINO IR that is compatible with Ollama. Here, we use the DeepSeek-R1-Distill-Qwen-7B model as an example:
1. Download the OpenVINO IR Model
Download the model from ModelScope:
pip install modelscope
modelscope download --model zhaohb/DeepSeek-R1-Distill-Qwen-7B-int4-ov --local_dir ./DeepSeek-R1-Distill-Qwen-7B-int4-ov2. Package the Downloaded OpenVINO IR Directory
Compress the directory into a *.tar.gz file:
tar -zcvf DeepSeek-R1-Distill-Qwen-7B-int4-ov.tar.gz DeepSeek-R1-Distill-Qwen-7B-int4-ov3. Create a Modelfile
Define the model configuration in a Modelfile:
FROM DeepSeek-R1-Distill-Qwen-7B-int4-ov.tar.gz
ModelType "OpenVINO"
InferDevice "GPU"
PARAMETER stop ""
PARAMETER stop "```"
PARAMETER stop "</User|>"
PARAMETER stop "<|end_of_sentence|>"
PARAMETER stop "</|"
PARAMETER max_new_token 4096
PARAMETER stop_id 151643
PARAMETER stop_id 151647
PARAMETER repeat_penalty 1.5
PARAMETER top_p 0.95
PARAMETER top_k 50
PARAMETER temperature 0.84. Create an Ollama-Compatible Model
Use the Modelfile to create a model supported by Ollama:
ollama create DeepSeek-R1-Distill-Qwen-7B-int4-ov:v1 -f Modelfile
With these steps, we have successfully created the DeepSeek-R1-Distill-Qwen-7B-int4-ov:v1 model, which is now ready for use with the Ollama OpenVINO backend.
OpenVINO toolkit for ARM platforms overview
OpenVINO, an advanced framework for neural network inference, has expanded its capabilities to include support for ARM architecture. Leveraging the streamlined and lightweight design of ARM processors, OpenVINO boosts its efficiency in AI tasks, which in turn widens application possibilities. To ensure a flawless experience within the OpenVINO ecosystem, the toolkit's CPU plugin has been refined for ARM architecture, focusing on improved performance and memory optimization, particularly for AI workloads running on ARM processors.
This article describes the ARM component within the CPU plugin, outlines the initiatives to back the development effort, and explains how it relates to the concept of OpenVINO toolkit. This article serves as an introduction and the first in a series of articles on ARM architecture support in OpenVINO.
ARM integrations in OpenVINO toolkit
Introduction to the ARM Integration within the OpenVINO CPU Plugin
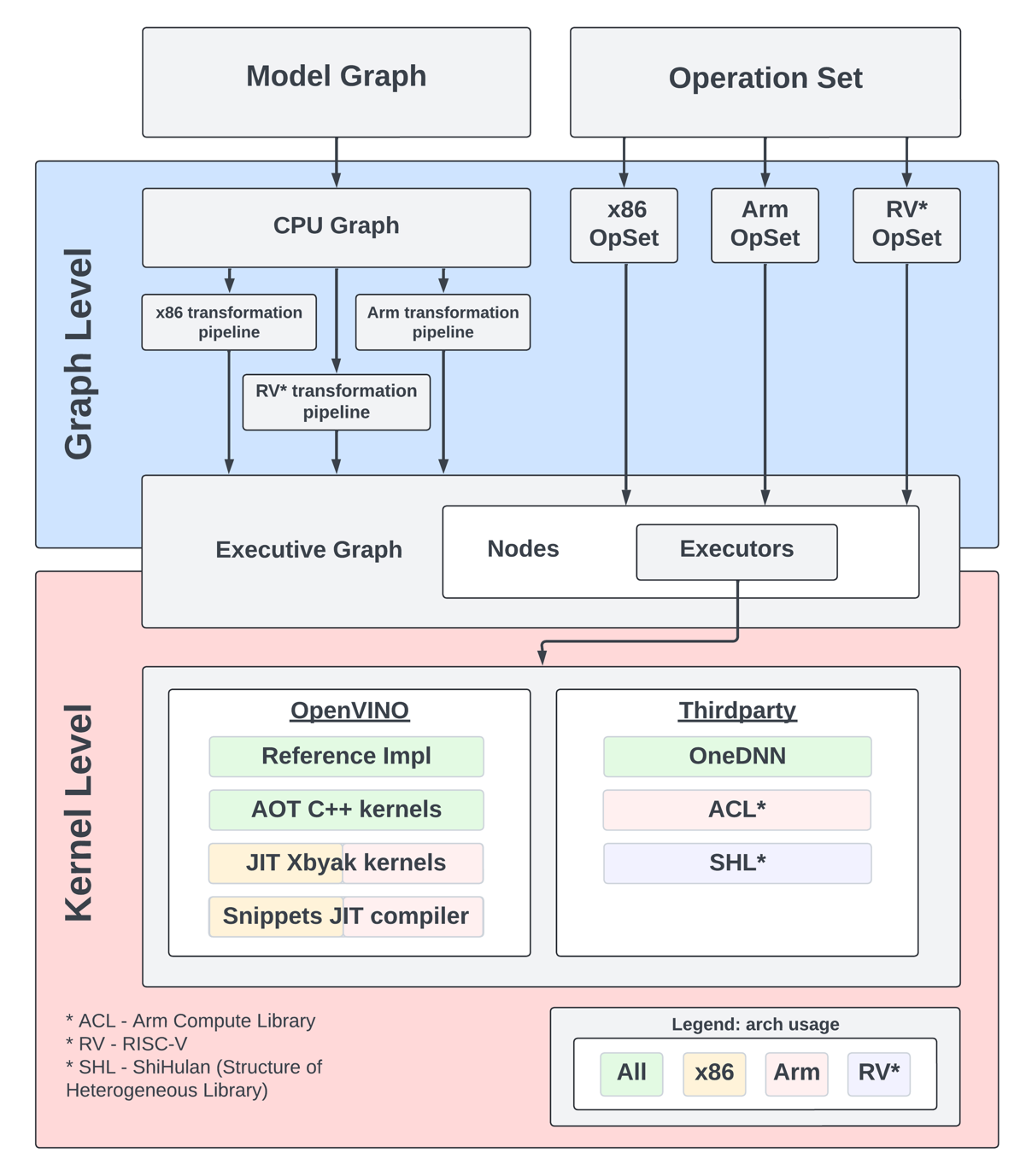
The OpenVINO CPU plugin was selected as the base for ARM architecture because of operational and architectural similarities between ARM and x86 platforms.
The OpenVINO CPU plugin architecture is divided into two main parts: the graph level and the execution kernel level, as shown in Diagram 1. The graph level involves optimizing the base graph of the AI model and converting it into an internal representation, using information about the processor architecture, computation specifics, algorithm characteristics, and more. The kernel level contains a set of computational kernels packaged into executors for various platforms. This set of computational kernels is divided into two main groups: kernels implemented directly within OpenVINO, and kernels utilized from third-party libraries. These groups help to select the optimal solutions for all supported architectures.
It is important to highlight that OpenVINO includes a special optimization layer called graph transformations. The main goal of this layer is to optimize the computation of the model graph on the processor, as well as to determine the order of processing and execution of graph nodes (or to eliminate some nodes from computations). Additionally, this layer has two significant aspects. The wide set of general transformations helps the plugin adjust to new architectures and keeps the computation graph well-suited for each specific type, like x86 or ARM. Balancing between general and specific optimizations, this approach is a crucial part of processing the model graph before execution by the kernels.
Generic Approaches for Device-Specific Optimizations in the CPU Plugin:
The CPU plugin incorporates three primary approaches to enable efficient, device-specific optimizations. These approaches are fully adopted for ARM architecture:
- Compiler Approach (Snippets)
- Integration ofOpenVINO Graph Compiler(known as Snippets)
The CPU plugin integrates Snippets to optimize performance by identifying and reusing common patterns within the execution graph. This approach enhances computational efficiency and ensures adaptability across different tasks.
- Integration ofOpenVINO Graph Compiler(known as Snippets)
- OpenVINO internal kernels (JIT + AOT):
- JIT (Xbyak): The CPU plugin employs JIT-compiled kernels using Xbyak to dynamically generate optimized code at runtime.
- AOT (Optimal Reference Implementation): Precompiled kernels developed within the OpenVINO framework provide efficient, reusable solutions for computational tasks.
- Third-Party Libraries
The CPU plugin leverages third-party libraries to access pre-optimized kernels for ARM:- Direct Use ofARM Compute Library(ACL) Kernels:
By directly using kernels from the ARM Compute Library (ACL), the plugin exploits the library’s inherent advantages for ARM-based computations. - Accessing ACL Kernels viaOneDNN:
The plugin accesses and optimizes ACL kernels through OneDNN, ensuring comprehensive coverage of ACL’s capabilities. - Accessing Other ARM-Optimized Kernels via OneDNN:
Beyond ACL, the plugin supports additional ARM-optimized kernels available through OneDNN, enabling broader model compatibility.
- Direct Use ofARM Compute Library(ACL) Kernels:
It is also important to address the topic of parallelism. In the context of OpenVINO, there are two levels of parallelism: graph-level parallelism and kernel-level parallelism. This approach enables optimal distribution of the processor load and maximizes the use of computational resources. Due to its complexity, a custom interface for parallelism is necessary, which incorporates particular libraries and standards for parallel computing. For ARM architecture support in OpenVINO, the current parallelism approach employs the OpenMP standard or the OneTBB library. OpenVINO uses OneTBB as the default threading backend, offering scalability and efficient task scheduling for high-performance applications. For users who prefer OpenMP, OpenVINO can be recompiled from source to enable OpenMP support. This flexibility allows developers to tailor threading options to specific project needs.For users who prefer OpenMP, OpenVINO can be recompiled from source to enable OpenMP support. This flexibility allows developers to tailor threading options to specific project needs.
Additionally, it is useful to briefly explain the role of external third-party libraries in OpenVINO.
To gain a deeper understanding of the context, we will discuss key libraries such as ARM Compute Library and OneDNN. These libraries are crucial for enhancing performance and reducing memory consumption in the OpenVINO CPU plugin for ARM architecture.
Arm Compute Library
The Arm Compute Library is an open-source collection of software optimized for Cortex-A CPUs, Neoverse systems, and Mali GPUs. It offers superior performance compared to other open-source alternatives and rapidly integrates new Arm technologies like SVE2. Key features include over 100 machine learning functions for CPU and GPU, support for multiple convolution algorithms (GEMM, Winograd, FFT, Direct), and various data types (FP32, FP16, int8, uint8, BFloat16). The library provides micro-architecture optimizations for key ML primitives, highly configurable build options for lightweight binaries, and advanced techniques such as Kernel Fusion, Fast math, and texture utilization. Additionally, it supports device and workload-specific tuning with OpenCL tuner and GEMM-optimized heuristics.
OneDNN Library
Intel® oneAPI Deep Neural Network Library (oneDNN) provides highly optimized implementations for deep learning operations across CPUs, GPUs, and other hardware. Its unified API improves performance for frameworks such as OpenVINO, Intel AI Tools, PyTorch*, and TensorFlow*, streamlining development and deployment processes by eliminating the need for target-specific code.
This library is fundamental to OpenVINO as it is crucial for optimizing neural network inference on Intel processors. It has drawn interest for supporting various computational architectures, made possible by the developer community and the accessibility of its source code. For instance, the ARM Compute Library is used for ARM architectures, along with JIT kernels integrated by Fujitsu. This variety enables achieving optimal performance on ARM architectures.
OpenVINO Distribution
The OpenVINO™ Runtime distribution for ARM devices provides various easy installation options. You can install OpenVINO via an archive file, use Python's PyPI, Conda Forge, Homebrew for macOS, Microsoft's vcpkg package manager, or Conan Package Manager. Detailed instructions are provided for each method to help you through the setup.
OpenVINO Notebooks
OpenVINO notebooks provide tutorials and step-by-step guides for various deep learning tasks. They cover a range of topics from setup process to model optimization and deployment. The notebooks are useful for learners at all levels, providing clear explanations and examples to help you learn OpenVINO.
The demos below demonstrate several notebooks running locally on Apple Mac M1 Pro Laptop and Apple Mac M2 Studio with OpenVINO™ 2025.0 release.
The Phi-3 Vision model is used for multimodal tasks, combining text and image processing. It can generate image descriptions, answer questions based on visual content, extract and analyze text from images, classify objects and scenes, and assist in content moderation.
https://drive.google.com/file/d/1-deK-heXq9B-iu4kAIO3ty8mDxLE3cnx/view?usp=sharing
Stable Diffusion 3 is a next-gen diffusion model using MMDiT, offering superior image quality, typography, and prompt adherence with improved efficiency.
https://drive.google.com/file/d/1HDB_d8sTkvqAstoq39QzufCF3rvYYjjJ/view?usp=sharing
Yolo-v11 object detection example
YOLOv11 is a fast and efficient real-time object detection model with improved accuracy and optimized architecture. It supports tasks like detection, segmentation, and classification while being adaptable to various platforms.
https://drive.google.com/file/d/1ISlBEAQWpyiEvFo6aRDBlQcfnf0l-V3R/view?usp=sharing
Collaborating with the Open-source Community
Good First Issue tasks
Developers eager to help improve OpenVINO can start by tackling "good first issue" tickets for ARM devices, which are a helpful way to enhance OpenVINO's performance on ARM architectures.
Contributing to the development of the OpenVINO plugin for ARM devices involves optimizing performance, aligning with ARM's features, and tackling platform-specific challenges. The open-source community is actively working on the plugin, taking on advanced tasks like implementation of highly optimized JIT kernels.
Google Summer of Code
Last year, our team joined the Google Summer of Code with a project aimed at boosting the performance of Generative AI (GenAI) on ARM devices using the OpenVINO toolkit.
The project focused on decreasing latency, accelerating model compilation, and reducing memory consumption. It entailed creating a benchmarking system and using advanced optimization methods for ARM architectures in the OpenVINO ecosystem. This work was important for OpenVINO's growth as an open-source framework.
Conclusion
In summary, using the OpenVINO CPU plugin on ARM devices can significantly improve computational efficiency and accelerate inference tasks. Its optimization techniques and compatibility with ARM architectures help developers make the most of ARM-based platforms for diverse AI applications. As ARM devices become more common in different industries, OpenVINO toolkit stands out as a powerful way to get fast AI results with lower latency and power consumption. Additionally, the teamwork between Intel and the ARM community is driving new developments in AI deployment for desktops and servers.
OpenVINO deploying DeepSeek-R1 Model Server (OVMS) on Bare metal Windows AIPC
Authors: Kunda Xu, Sapala, Rafal A
DeepSeek-R1 is an open-source reasoning model developed by DeepSeek to address tasks requiring logical inference, mathematical problem-solving, and real-time decision-making. With DeepSeek-R1,you can follow its logic, making it easier to understand and, if necessary, challenge its output. This capability gives reasoning models an edge in fields where outcomes need to be explainable, like research or complex decision-making.
Distillation in AI creates smaller, more efficient models from larger ones, preserving much of their reasoning power while reducing computational demands. DeepSeek applied this technique to create a suite of distilled models from R1, using Qwen and Llama architectures. That allows us to try DeepSeek-R1 capability locally on usual laptops (AIPC).
In this tutorial, we consider deploy deepseek-ai/DeepSeek-R1-Distill-Qwen-7B as a model server on Intel AIPC or AI work station with Windows OS to perform request generation tasks.
Requirements:
- Windows11
- VC_redist : Microsoft Visual C++ Redistributable
- Intel iGPU or ARC GPU: Intel® Arc™ & Iris® Xe Graphics - Windows*
- OpenVINO >=2025.0 : https://github.com/openvinotoolkit/openvino/releases/tag/2025.0.0
- OVMS >=2025.0 : https://github.com/openvinotoolkit/model_server/releases/tag/v2025.0
QuickStart Guide
Step 1. Install python dependencies for the conversion script:
Step 2. Run optimum-cli to download and quantize the model:
If your network access to HuggingFace is unstable, you can try to use a proxy image to pull the model.
Step 3. Deploying Model Server (OVMS) on Bare metal
Download and unpack model server archive for Windows:
Run setupvars script to set required environment variables
Step 4. DeepSeek-R1 model server deploy
Bare metal Host deploy. Required: deploying ovms on Bera metal.
OpenVINO + OVMS can also use Docker contain deploying. Required: Docker engine installed
When using docker as a deployment method, you need to consider whether the hardware performance of the machine is sufficient, because docker contain will also generate additional memory overhead.
For example, when deploying on a laptop or AIPC, due to the limited memory resources, it is more reasonable to use bare metal deployment method
Step 5. Check readiness Wait for the model to load.
You can check the status with a simple command
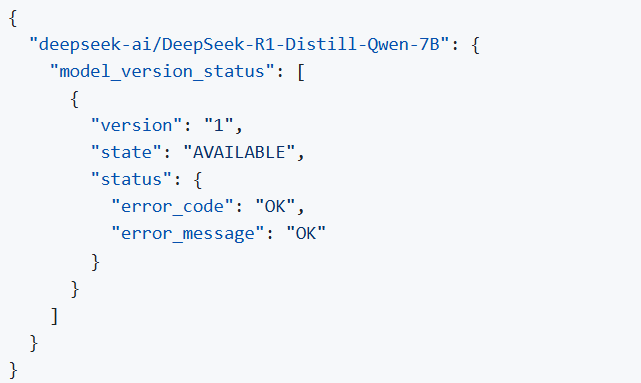
Step 6. Run model server generation
Create a file called request.json ,
and copy the following content into it
You will get the output like the following.

Note: If you want to get the response chunks streamed back as they are generated change stream parameter in the request to true
DeepSeek Janus-Pro Model Enabling with OpenVINO
1. Introduction
Janus is a unified multimodal understanding and generation model developed by DeepSeek. Janus proposed decoupling visual encoding to alleviate the conflict between multimodal understanding and generation tasks. Janus-Pro further scales up the Janus model to larger model size (deepseek-ai/Janus-Pro-1B & deepseek-ai/Janus-Pro-7B) with optimized training strategy and training data, achieving significant advancements in both multimodal understanding and text-to-image tasks.
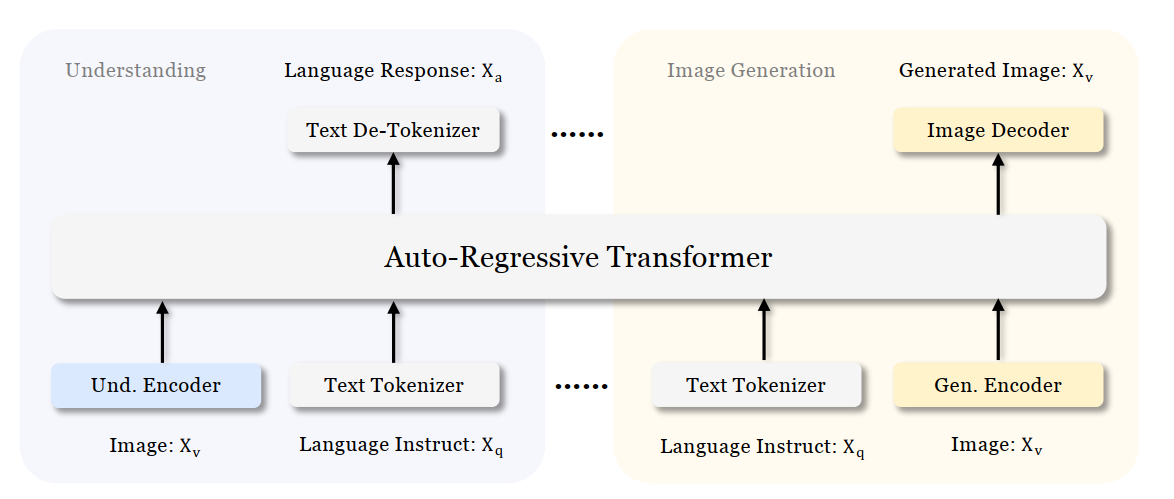
Figure 1 shows the architecture of Janus-Pro, which decouples visual encoding for multimodal understanding and visual generation. “Und. Encoder” and “Gen. Encoder” are abbreviations for “Understanding Encoder” and “Generation Encoder”. For the multimodal understanding task, SigLIP vision encoder used to extract high-dimensional semantic features from the image, while for the vision generation task, VQ tokenizer used to map images to discrete IDs. Both the understanding adaptor and the generation adaptor are two-layer MLPs to map the embeddings to the input space of LLM.
In this blog, we will introduce how to deploy Janus-Pro model with OpenVINOTM runtime on the intel platform.
2. Janus-Pro Pytorch Model to OpenVINOTM Model Conversion
2.1. Setup Python Environment
2.2 Download Janus Pytorch model (Optional)
2.3. Convert Pytorch Model to OpenVINOTM INT4 Model
The converted OpenVINO will be saved in Janus-Pro-1B-OV directory for deployment.
3. Janus-Pro Inference with OpenVINOTM Demo
In this section, we provide several examples to show Janus-Pro for multimodal understanding and vision generation tasks.
3.1. Multimodal Understanding Task – Image Caption with OpenVINOTM
Prompt: Describe image in details
Input image:

Generated Output:
3.2. Multimodal Understanding Task – Equation Description with OpenVINOTM
Prompt: Generate the latex code of this formula
Input Image:
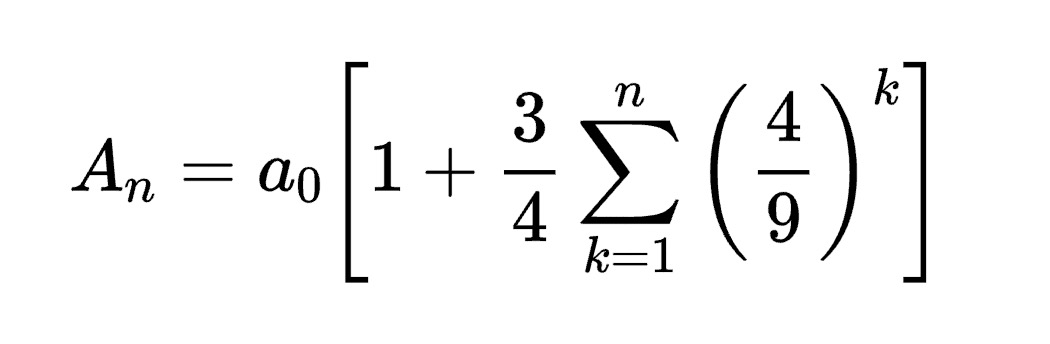
Generated Output:
3.3. Multimodal Understanding Task – Code Generation with OpenVINOTM
Prompt: Generate the matplotlib pyplot code for this plot
Input Image:
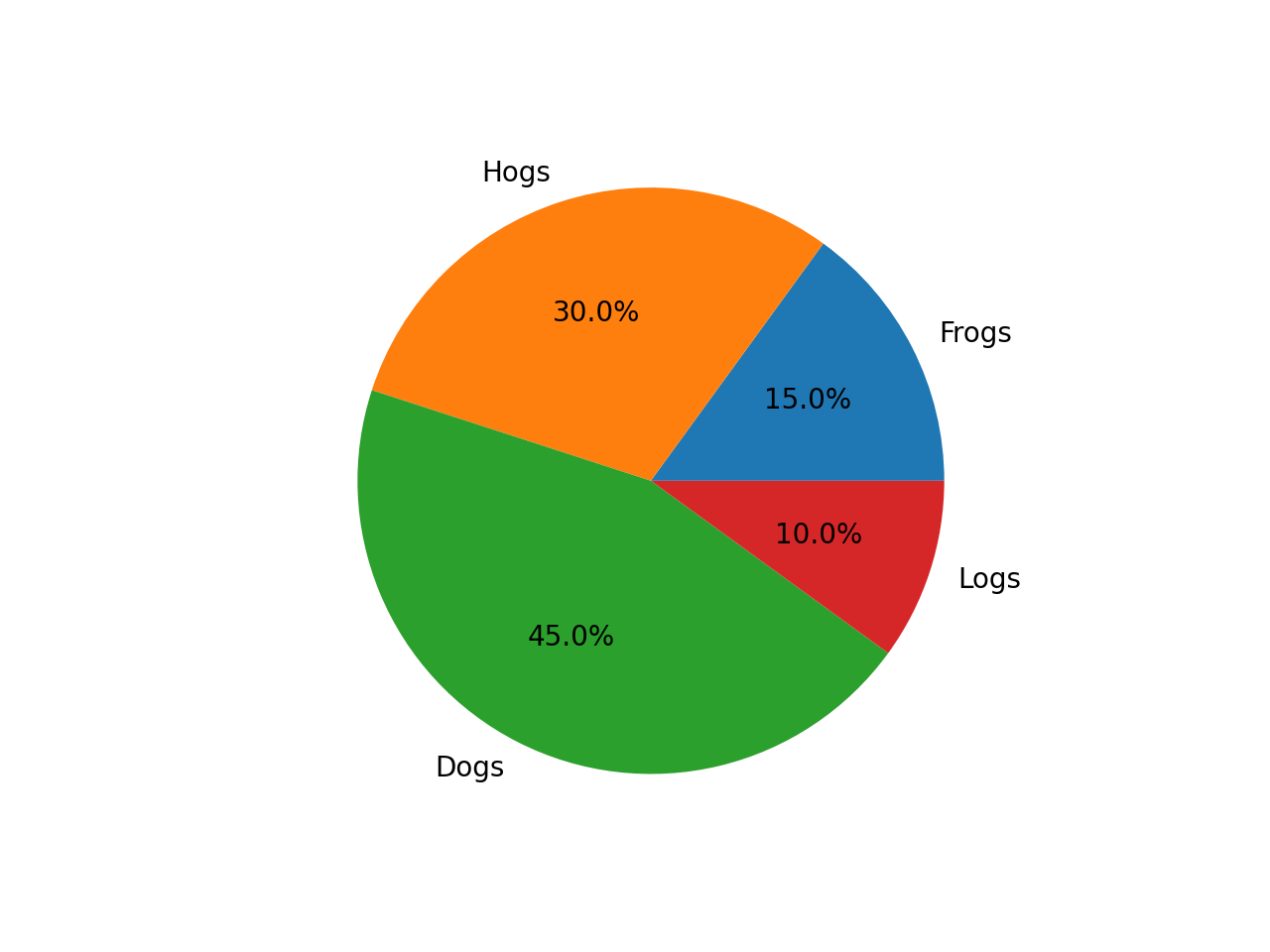
Generated Output:
3.4. Vision Generation Task with OpenVINOTM
Input Prompt: A cute and adorable baby fox with big brown eyes, autumn leaves in the background enchanting, immortal, fluffy, shiny mane, Petals, fairyism, unreal engine 5 and Octane Render, highly detailed, photorealistic, cinematic, natural colors.
Generated image:


4. Performance Evaluation & Memory Usage Analysis
We also provide benchmark scripts to evaluate Janus-Pro model performance and memory usage with OpenVINOTM inference, you may specify model name and device for your target platform.
4.1. Benchmark Janus-Pro for Multimodal Understanding Task with OpenVINOTM
Here are some arguments for benchmark script for Multimodal Understanding Task:
--model_id: specify the Janus OpenVINOTM model directory
--prompt: specify input prompt for multimodal understanding task
--image_path: specify input image for multimodal understanding task
--niter: specify number of test iteration, default is 5
--device: specify which device to run inference
--max_new_tokens: specify max number of generated tokens
By default, the benchmark script will run 5 round multimodal understanding tasks on target device, then report pipeline initialization time, average first token latency (including preprocessing), 2nd+ token throughput and max RSS memory usage.
4.2. Benchmark Janus-Pro for Text-to-Image Task with OpenVINOTM
Here are some arguments for benchmark scripts for Text-to-Image Task
--model_id: specify the Janus OpenVINO TM model directory
--prompt: specify input prompt for text-to-image generation task
--niter: specify number of test iteration
--device: specify which device to run inference
By default, the benchmark script will run 5 round image generation tasks on target device, then report the pipeline initialization time, average image generation latency and max RSS memory usage.
5. Conclusion
In this blog, we introduced how to enable Janus-Pro model with OpenVINOTM runtime, then we demonstrated the Janus-Pro capability for various multimodal understanding and image generation tasks. In the end, we provide python script for performance & memory usage evaluation for both multimodal understanding and image generation task on target platform.
Q4'24: Technology Update – Low Precision and Model Optimization
Authors
Alexander Kozlov, Nikolay Lyalyushkin, Nikita Savelyev, Souvikk Kundu, Andrey Anufriev, Pablo Munoz, Alexander Suslov, Liubov Talamanova, Daniil Lyakhov, Yury Gorbachev, Nilesh Jain, Maxim Proshin
Summary
What a quarter! Tons of works for Transformer model optimization in Q4’24 including fundamental ones such as “scaling lows for quantized LLMs“. Such a huge effort can indicate a growing adoption of LLMs and AI in general and the need for a further cost reduction. We had to extend the Highlights to six papers this time considering the amount of work being done.
Highlights
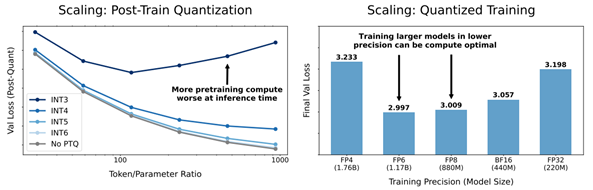
- Scaling Laws for Precision by Harvard, Stanford, MIT, Carnegie Mellon Universities, and Databricks (https://arxiv.org/pdf/2411.04330). In this work, authors devise “precision-aware” scaling laws for both training and inference. They propose that training in lower precision reduces the model’s effective parameter count, allowing predicting the additional loss incurred from training in low precision and post-train quantization. For inference, they find that the degradation introduced by post-training quantization increases as models are trained on more data, eventually making additional pretraining data actively harmful. For training, their scaling laws allow predicting the loss of a model with different parts in different precisions and suggest that training larger models in lower precision may be compute optimal. Authors unify the scaling laws for post and pretraining quantization to arrive at a single functional form that predicts degradation from training and inference in varied precisions. They fit on over 465 pretraining runs and validate our predictions on model sizes up to 1.7B parameters trained on up to 26B tokens.
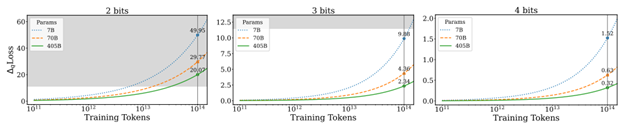
- Low-Bit Quantization Favors Undertrained LLMs: Scaling Laws for Quantized LLMs with100T Training Tokens by University of Virginia, Tencent AI Lab Seattle (https://arxiv.org/pdf/2411.17691).Authors propose a perspective that one can use to measure an LLM’s training levels and determine the number of training tokens required for fully training LLMs of various sizes. Moreover, authors use the scaling laws to predict the quantization performance of different-sized LLMs trained with 100 trillion tokens. Our projection shows that the low-bit quantization performance of future models, which are expected to be trained with over 100 trillion tokens, may NOT be desirable. This poses a potential challenge for low-bit quantization in the future and highlights the need for awareness of a model’s training level when evaluating low-bit quantization research. Checkpoints are available at: https://huggingface.co/Xu-Ouyang.
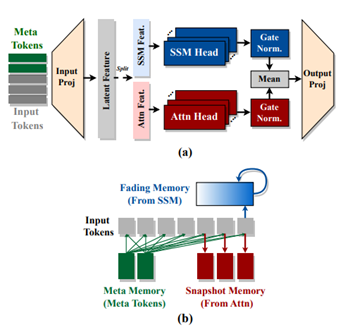
- Hymba: A Hybrid-head Architecture for Small Language Models by Nvidia, Georgia Institute of Technology, and HKUST (https://www.arxiv.org/abs/2411.13676).The paper introduces a family of small language models featuring a hybrid-head parallel architecture that integrates transformer attention mechanisms with state space models (SSMs) for enhanced efficiency. Additionally, authors introduce learnable meta tokens that are prepended to prompts, storing critical information. This model is further optimized by incorporating cross-layer key-value (KV) sharing and partial sliding window attention, resulting in a compact cache size. Hymba-1.5B-Base model surpasses all sub-2B public models in performance and even outperforms Llama-3.2-3B with1.32% higher average accuracy, an 11.67× cache size reduction, and 3.49×throughput. Models are available on the Hugging Face Hub.
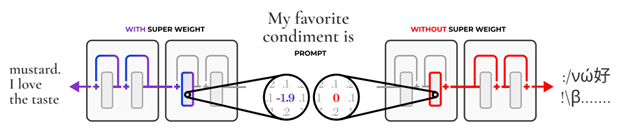
- THE SUPER WEIGHT IN LARGE LANGUAGE MODELS by Apple and University of Notre Dame (https://arxiv.org/pdf/2411.07191). This work presents a finding that pruning single parameters can destroy an LLM’s ability to generate text – increasing perplexity by 3 orders of magnitude and reducing zero-shot accuracy to guessing. It proposes a data-free method for identifying such parameters, termed super weights, using a single forward pass through the model. Authors find that these super weights induce correspondingly rare and large activation outliers, termed super activations. When preserved with high precision, super activations can improve simple round-to-nearest quantization to become competitive with state-of-the-art methods. For weight quantization, they similarly find that by preserving the super weight and clipping other weight outliers, round-to-nearest quantization can scale to much larger block sizes than previously considered. The code is available at n https://github.com/mengxiayu/LLMSuperWeight.
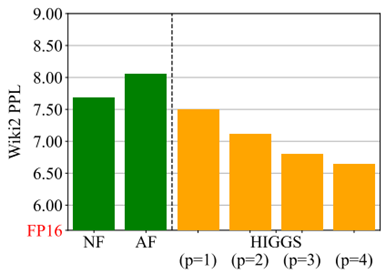
- Pushing the Limits of Large Language Model Quantization via the Linearity Theorem by Yandex, HSE University, ISTA, GenAI CoE, KAUST, Neural Magic (https://arxiv.org/pdf/2411.17525). The paper presents a “linearity theorem” establishing a direct relationship between the layer-wise ℓ2 reconstruction error and the model perplexity increase due to quantization. This enables two novel applications: (1) a simple data-free LLM quantization method using Hadamard rotations and MSE-optimal grids, dubbed HIGGS, which outperforms all prior data-free approaches such as the extremely popular NF4 quantized format, and (2) an optimal solution to the problem of finding non-uniform per-layer quantization levels which match a given compression constraint in the medium-bit width regime, obtained by reduction to dynamic programming. Authors demonstrate improved accuracy-compression trade-offs on Llama-3.1 and 3.2- family models, as well as on Qwen-family models.
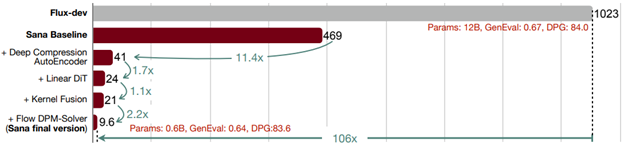
- SANA:EFFICIENT HIGH-RESOLUTION IMAGE SYNTHESIS WITH LINEAR DIFFUSION TRANSFORMERS by NVIDIA, MIT, Tsinghua University (https://arxiv.org/pdf/2410.10629). Authors introduce Sana, a text-to-image frame work that can generate images up to 4096×4096 resolution. Core designs include: (1) Deep compression autoencoder: unlike traditional AEs, which compress images only 8×,authors trained an AE that can compress images 32×, effectively reducing the number of latent tokens. (2) Linear DiT: they replace all vanilla attention in DiT with linear attention (3) Decoder-only text encoder: they replaced T5 with modern decoder-only small LLM as the text encoder and designed complex human instruction with in-context learning to enhance the image-text alignment. (4) Efficient training and sampling: they propose Flow-DPM-Solver to reduce sampling steps. As a result, Sana-0.6B is very competitive with modern giant diffusion model (e.g. Flux-12B), being 20times smaller and 100+ times faster in measured throughput. Project web page with code: https://nvlabs.github.io/Sana/.
Papers with notable results
Quantization
- VPTQ: EXTREME LOW-BIT VECTOR POST-TRAINING QUANTIZATION FOR LARGE LANGUAGE MODELS by Microsoft and University of Science and Technology of China (https://arxiv.org/abs/2409.17066). The authors introduce Vector Post-Training Quantization and use Second-Order Optimization to formulate the LLM VQ problem and guide the algorithm design by solving the optimization. They further refine the weights using Channel-Independent Second-Order Optimization for a granular VQ. In addition, by decomposing the optimization problem, authors propose a brief codebook initialization algorithm and extend VPTQ to support residual and outlier quantization, which enhances model accuracy and further compresses the model. The method achieves good results on llama-2 and llama-3 model families, resulting in a 1.6-1.8× increase in inference throughput compared to SOTA. The code is available at https://github.com/microsoft/VPTQ.
- ADDITION IS ALL YOU NEED FOR ENERGY-EFFICIENT LANGUAGE MODELS by BitEnergy AI (https://arxiv.org/pdf/2410.00907). Authors propose the linear-complexity multiplication algorithm that approximates floating point number multiplication with integer addition operations. The new algorithm costs significantly less computation resource than 8-bit floating point multiplication but achieves higher precision. Compared to 8-bit floating point multiplications, the proposed method achieves higher precision but consumes significantly less bit-level computation which can potentially reduce 95% energy cost by elementwise floating point tensor multiplications and 80% energy cost of dot products. A numerical analysis and experiments indicate that the method with 4-bit mantissa achieves comparable precision as float8 e4m3 multiplications, and with 3-bit mantissa outperforms float8 e5m2. Evaluation results on popular benchmarks show that directly applying L-Mul to the attention mechanism is almost lossless.
- BitNet a4.8: 4-bit Activations for 1-bit LLMs by Microsoft and University of Chinese Academy of Sciences (https://arxiv.org/pdf/2411.04965). In this work, authots introduce BitNet a4.8, enabling 4-bit activations for 1-bit LLMs. BitNet a4.8 employs a hybrid quantization and sparsification strategy to mitigate the quantization errors introduced by the outlier channels. Specifically, they utilize 4-bit activations for inputs to the attention and feed-forward network layers, while sparsifying intermediate states followed with 8-bit quantization. Extensive experiments demonstrate that BitNet a4.8 achieves performance comparable to BitNet b1.58 with equivalent training costs, while being faster in inference with enabling 4-bit (INT4/FP4) kernels. Additionally, BitNet a4.8 activates only 55% of parameters and supports 3-bit KV cache.
- MagR: Weight Magnitude Reduction for Enhancing Post-Training Quantization by Uniiversity at Albany and IBM (https://arxiv.org/pdf/2406.00800). MagR is an optimization-based preprocessing technique for improving post-training quantization. It solves an l_∞-regularized problem to reduce outlier weights and center them around zero, enabling smoother and more efficient quantization. Unlike linear transformations that require extra steps at inference, MagR is a non-linear transformation that adds no overhead. Experiments show state-of-the-art results, including a Wikitext2 perplexity of 6.7 on the LLaMA2-70B model using per-channel INT2 weight quantization.
- Cherry on Top: Parameter Heterogeneity and Quantization in Large Language Models by Shanghai University of Finance and Economics (https://arxiv.org/pdf/2404.02837). This paper identifies “cherry” parameters in large language models—those few parameters with a disproportionately large effect on performance—while most parameters matter far less. Building on this insight, the authors introduce CherryQ, a quantization technique that maintains these critical parameters in high precision and aggressively quantizes the rest. CherryQ delivers improved perplexity and downstream task results, enabling efficient LLM deployment. Remarkably, a 3-bit quantized Vicuna-1.5 model matches the performance of 16-bit models, illustrating the potential of leveraging parameter heterogeneity for more efficient inference.
- QTIP: Quantization with Trellises and Incoherence Processing by Cornell University (https://arxiv.org/pdf/2406.11235). QTIP is a new PTQ approach leveraging trellis-coded quantization (TCQ) for ultra-high-dimensional vector quantization of LLM weights. Unlike conventional VQ methods whose codebook size grows exponentially with dimension, TCQ uses a stateful decoder to maintain efficiency as dimensions scale. QTIP provides a hardware-friendly “bitshift” trellis structure and can be tuned for lookup-only or computed lookup-free decoding. This allows faster, more memory-efficient inference and achieves state-of-the-art quantization quality, outperforming previous VQ-based methods.
- ESPACE: Dimensionality Reduction of Activations for Model Compression by NVIDIA (https://arxiv.org/pdf/2410.05437). ESPACE introduces a new LLM compression method based on dimensionality reduction of activations rather than weight decomposition. By projecting activations onto pre-calibrated principal components, ESPACE retains model expressivity without retraining. It achieves weight compression indirectly through matrix multiplication associativity. Theoretically, it ensures optimal computational accuracy when constructing projection matrices. Experiments show up to 50% compression on GPT3, Llama2, and Nemotron4 with minimal accuracy loss, and in some cases, improved perplexity. ESPACE also speeds up inference. Compared to existing tensor decomposition methods, ESPACE advances state-of-the-art LLM compression.
- Delta-CoMe: Training-Free Delta-Compression with Mixed-Precision for Large Language Models by several Chinese universities (https://arxiv.org/pdf/2406.08903). This work addresses compressing delta weights for fine-tuned LLMs, where maintaining task-specific performance is challenging using low-rank or low-bit methods. Observing that delta weights’ singular values are long-tailed, the authors propose a mixed-precision delta quantization approach. By assigning higher-bit precision to more influential singular vectors, their method preserves accuracy. Experiments on diverse fine-tuned LLMs—including math, code, and chat models—show that this approach matches full-precision performance and significantly outperforms standard low-rank and low-bit baselines. It is also compatible with various backbone models, such as Llama-2, Llama-3, and Mistral.
- StepbaQ: Stepping backward as Correction for Quantized Diffusion Models by MediaTek and Purdue University (https://openreview.net/pdf?id=cEtExbAKYV). StepbaQ reframes quantization error in diffusion models as a “stepback” in their denoising process. By analyzing how this accumulated error distorts the sampling trajectory, StepbaQ introduces a correction mechanism that uses quantization error statistics from a small calibration dataset. Without altering quantization settings, it significantly improves model quality. For instance, StepbaQ boosts the FID score of quantized SD v1.5 by 7.30 under W8A8, and SDXL-Turbo by 17.31 under W4A8. This plug-and-play solution enhances performance on resource-constrained devices while maintaining broad applicability.
- LLMCBench: Benchmarking Large Language Model Compression for Efficient Deployment by Beihang University, ETH Zurich and Canerige Mellon University (https://arxiv.org/pdf/2410.21352). LLMCBench is a comprehensive benchmark designed to evaluate large language model compression techniques under realistic conditions. Moving beyond limited and specialized assessments, it tests various models, datasets, and metrics. LLMCBench establishes clearly defined evaluation tracks based on real production requirements and conducts extensive experiments with multiple mainstream compression methods. Through in-depth analysis, it offers insights into the strengths and weaknesses of these approaches. Ultimately, LLMCBench aims to guide the selection and design of effective compression algorithms, serving as a valuable resource for future research and development in LLM efficiency.
- DuQuant: Distributing Outliers via Dual Transformation Makes Stronger Quantized LLMs (https://duquant.github.io/). Generalization of the SmoothQuant algorithm which allows to mitigate the massive outliers and quantize not just LLM weights but activations as well. Shows promising results for LLama2/3 -8B W6A6 and W4A4 quantization. The code is available at: https://github.com/Hsu1023/DuQuant.
- Efficient Multi-task LLM Quantization and Serving for Multiple LoRA Adapters (https://openreview.net/pdf?id=HfpV6u0kbX). Multi quantized Lora adapters quantization via techniques like Multi-Lora GPTQ and LoRa Inlaid. Technics to dynamically add a new task/dataset to existing quantized LLM are discussed in the paper, promising pipeline for quantized LLM serving / update is presented.
- PROGRESSIVE MIXED-PRECISION DECODING FOR EFFICIENT LLM INFERENCE. Samsung AI Center, Cambridge UK, Imperial College London UK (https://arxiv.org/abs/2410.13461). The authors propose a novel phase-aware method that selectively allocates precision during different phases of LLM inference, achieving both strong context extraction during prefill and efficient memory bandwidth utilization during decoding. To further address the memory-boundedness of the decoding phase, the authors introduce Progressive Mixed-Precision Decoding (PMPD), a technique that enables the gradual lowering of precision deeper in the generated sequence, together with a spectrum of precision-switching schedulers that dynamically drive the precision lowering decisions in either task-adaptive or prompt-adaptive manner. Extensive evaluation across diverse language tasks shows that when targeting Nvidia GPUs, PMPD achieves 1.4−12.2× speedup in LLM linear layers over fp16 models, while when targeting an LLM-optimized NPU, our approach delivers a throughput gain of 3.8−8.0× over fp16 models and up to 1.54× over uniform quantization approaches while preserving the output quality.
- AMXFP4: TAMING ACTIVATION OUTLIERS WITH ASYMMETRIC MICROSCALING FLOATING-POINT FOR 4-BIT LLM INFERENCE by Hanyang University and Rebellions Inc. (https://arxiv.org/pdf/2411.09909). Authors propose Asymmetric Microscaling 4-bit Floating-Point (AMXFP4) for efficient LLM inference. This data format leverages asymmetric shared scales to mitigate outliers while naturally capturing the asymmetry introduced by group-wise quantization. Unlike conventional 4-bit quantization methods that rely on data rotation and costly calibration, AMXFP4 uses asymmetric shared scales for direct 4-bit casting, achieving better quantization accuracy across various LLM tasks, including multi-turn conversations, long-context reasoning, and visual question answering The code is available at https://github.com/aiha-lab/MX-QLLM.git.
- SageAttention2 Technical Report: Accurate 4 Bit Attention for Plug-and-play Inference Acceleration by Tsinghua University (https://arxiv.org/pdf/2411.10958). Authors propose an improvement over the previous version of SageAttention method which utilizes 4-bit matrix multiplication (Matmul) alongside additional precision-enhancing techniques. First, they propose to quantize matrixes (Q, K) to INT4 in a warp-level granularity and quantize matrixes to FP8. Second, they propose a method to smooth Q and V, enhancing the accuracy of attention. Third, they propose an adaptive quantization method to ensure the end-to-end metrics over various models. Authors claim a good performance improvement at small drop of accuracy for large language processing, image generation, and video generation. The codes are available at https://github.com/thu-ml/SageAttention.
- CATASTROPHIC FAILURE OF LLM UNLEARNING VIA QUANTIZATION (https://openreview.net/pdf?id=lHSeDYamnz). The paper reveals that applying quantization to models that have undergone unlearning can restore the "forgotten" information. Authors conduct experiments using various quantization techniques across multiple precision levels to evaluate this phenomenon. They find that for unlearning methods with utility constraints, the unlearned model retains an average of 21% of the intended forgotten knowledge in full precision, which significantly increases to 83% after 4-bit quantization. They also provide a theoretical explanation for the observed phenomenon and propose a quantization-robust unlearning strategy aimed at mitigating this intricate issue. Results highlight a fundamental tension between preserving the utility of the unlearned model and preventing knowledge recovery through quantization, emphasizing the challenge of balancing these two objectives. The code is available at: https://anonymous.4open.science/r/FailureUnlearning-20DE.
- Llama Guard 3-1B-INT4: Compact and Efficient Safeguard for Human-AI Conversations by Meta (https://arxiv.org/pdf/2411.17713). Author used a complex approach to optimize Llama Guard 3-1B for mobile platforms. Namely, they reduce the number of decoder blocks and MLP width of Llama Guard 3-1B-INT4 using a block-level and neuron-level sensitivity analysis, respectively. They use quantization-aware training (QAT) to reduce the weight bitwidth to 4 and the activation bitwidth to 8, such that the model size is cut down by 4× and the model can be efficiently run via ExecuTorch’s XNNPACK backend. They make use of the fact that Llama Guard models only require a limited output vocabulary and reduce the unembedding layer output shape from 128k to 20. Finally, the authors fine-tune the model with distillation from a Llama Guard 2-8B teacher to recover any lost model quality resulting from the compression steps.
- MPQ-DM: Mixed Precision Quantization for Extremely Low Bit Diffusion Models by Institute of Computing Technology, University of Chinese Academy of Sciences, ETH Zurich, Beijing Jiaotong University (https://arxiv.org/pdf/2412.11549). The paper presents a Mixed-Precision Quantization method for Diffusion Models. It mainly relies on two techniques: (1) To mitigate the quantization error caused by outlier severe weight channels, authors propose an Outlier-Driven Mixed Quantization (OMQ) technique that uses Kurtosis to quantify outlier salient channels and apply optimized intra-layer mixed-precision bit-width allocation to recover accuracy performance within target efficiency. (2) To robustly learn representations crossing time steps, they construct a Time-Smoothed Relation Distillation (TRD) scheme between the quantized diffusion model and its full-precision counterpart, transferring discrete and continuous latent to a unified relation space to reduce the representation inconsistency. The method achieves good generation results on public benchmarks in low-bit quantization settings, e.g. W3A6, W3A4. Code is planned to be released here.
- Panacea: Novel DNN Accelerator using Accuracy-Preserving Asymmetric Quantization and Energy-Saving Bit-Slice Sparsity by POSTECH, University of Michigan (https://arxiv.org/pdf/2412.10059). The paper discloses how to build AI accelerator that leverages Bit-Slice Sparsity for the most prominent integer quantization scheme W-sym, A-asym. In contrast to the previous bit-slice computing, the accelerator compresses frequent nonzero slices, generated by asymmetric quantization, and skips their operations. To increase the slice level sparsity of activations, authors also introduce two algorithm hardware co-optimization methods: a zero-point manipulation and a distribution-based bit-slicing.
- Efficiency Meets Fidelity: A Novel Quantization Framework for Stable Diffusion by Zhejiang University and vivo Mobile Communication Co (https://arxiv.org/pdf/2412.06661). The paper introduces a mix-precision quantization strategy, multi-timestep activation quantization, and time information precalculation techniques to ensure high fidelity image generation of Stable Diffusion models in comparison to floating-point counterparts. The method achieves a good consistency of the image generation under the W8A8 and W4A8 settings.
- PREFIXQUANT: STATIC QUANTIZATION BEATS DYNAMIC THROUGH PREFIXED OUTLIERS IN LLMS by The University of Hong Kong, Shanghai AI Laboratory, Tongji University (https://arxiv.org/pdf/2410.05265). The paper proposes a technique that isolates outlier tokens offline without re-training. Specifically, it identifies high-frequency outlier tokens and prefixes them in the KV cache, preventing the generation of outlier tokens during inference and simplifying quantization. The method achieves very promising results in LLM static quantizaiton. For instance, in W4A4KV4 Llama-3-8B, with per-tensor static quantization it achieves a 7.43 WikiText2 perplexity and 71.08% average accuracy on 5 common-sense reasoning tasks. Additionally, the inference speed of W4A4 quantized models using PrefixQuant is 1.60× to 2.81× faster than FP16. The code is available at https://github.com/ChenMnZ/PrefixQuant.
- MixPE: Quantization and Hardware Co-design for Efficient LLM Inference by The Chinese University of Hong, Tsinghua University, Huawei Noah’s Ark Lab (https://arxiv.org/pdf/2411.16158). The paper proposes performing dequantization after per-group mixed-precision GEMM, significantly reducing dequantization overhead. Second, instead of relying on conventional multipliers, the method utilizes efficient shift&add operations for multiplication, optimizing both computation and energy efficiency. Experimental results demonstrate that the proposed design achieves better performance and energy trade-offs.
- “GIVE ME BF16 OR GIVE ME DEATH”? ACCURACY-PERFORMANCE TRADE-OFFS IN LLM QUANTIZATION by Neural Magic, Institute of Science and Technology Austria (https://arxiv.org/pdf/2411.02355). A thorough investigation, encompassing over 500,000 individual evaluations, yields several key findings: (1) FP8 weight and activation quantization (W8A8-FP) is lossless across all model scales, (2) INT8 weight and activation quantization (W8A8-INT) incurs surprisingly low 1-3% accuracy degradation, and (3) INT4 weight-only quantization (W4A16-INT) is competitive with 8-bit integer weight and activation quantization. They find that W4A16 offers the best cost-efficiency for synchronous deployments and for asynchronous deployment on mid-tier GPUs. At the same time, W8A8 formats excel in asynchronous “continuous batching” deployment of mid- and large-size models on high-end GPUs.
- GWQ: Gradient-Aware Weight Quantization for Large Language Models by PKU, CASIA, THU, USTB, UNITN, ETHz, PolyU, UCAS (https://arxiv.org/pdf/2411.00850). The authors propose gradient-aware weight quantization that leverages gradients to localize outliers, requiring only a minimal amount of calibration data for outlier detection. It retains the weights corresponding to the top 1% outliers preferentially at FP16 precision, while the remaining non-outlier weights are stored in a low-bit format. GWQ found experimentally that utilizing the sensitive weights in the gradient localization model is more scientific than utilizing the sensitive weights in the Hessian matrix localization model. The method shows accurate results for both LLM and VLM quantization.
- SDP4Bit: Toward 4-bit Communication Quantization in Sharded Data Parallelism for LLM Training by Indiana University, ByteDance, and University of Houston (https://arxiv.org/pdf/2410.15526). The paper proposes a method that reduces the communication of weights and gradients during the training to nearly 4 bits via two techniques: quantization on weight differences, and two-level gradient smooth quantization. Furthermore, the method presents an algorithm system co-design with runtime optimization to minimize the computation overhead of compression. Authors empirically evaluate the accuracy on the pre-training of GPT models with up to 6.7 billion parameters, and the results demonstrate a negligible impact on training loss. Furthermore, speed experiments show up to 4.08× speedup in end-to-end throughput on a scale of 128 GPUs.
- Quamba: A Post-Training Quantization Recipe for Selective State Space Models by University of Texas at Austin, National Yang Ming Chiao Tung University, and Cornell University (https://arxiv.org/pdf/2410.13229). Authors propose a static 8-bit per-tensor SSM quantization method which suppresses the maximum values of the input activations to the selective SSM for finer quantization precision and quantizes the output activations in an outlier-free space with Hadamard transform. 8-bit weight-activation quantized Mamba 2.8B SSM benefits from hardware acceleration and achieves a 1.72 × lower generation latency on an Nvidia Orin Nano 8G, with only a 0.9% drop in average accuracy on zero-shot tasks. Code is released at https://github.com/enyac-group/Quamba.
- RESTRUCTURING VECTOR QUANTIZATION WITH THE ROTATION TRICK by Stanford University and Google DeepMind (https://arxiv.org/pdf/2410.06424). The paper proposes a way to propagate gradients through the vector quantization layer of VQ-VAEs. The method smoothly transforms each encoder output into its corresponding codebook vector via a rotation and rescaling linear transformation that is treated as a constant during backpropagation. As a result, the relative magnitude and angle between encoder output and codebook vector becomes encoded into the gradient as it propagates through the vector quantization layer and back to the encoder. Еhis restructuring improves reconstruction metrics, codebook utilization, and quantization error. Code is available at https://github.com/cfifty/rotation_trick.
Pruning / Sparsity
- MaskLLM: Learnable Semi-Structured Sparsity for Large Language Models by NVIDIA National University of Singapore (https://arxiv.org/pdf/2409.17481). The paper introduces several fundamental findings on applying N:M sparsity to LLM models. It explicitly models N:M patterns as a learnable distribution through Gumbel Softmax sampling. This approach facilitates end-to-end training on large-scale datasets and offers two notable advantages: 1) High-quality Masks - our method effectively scales to large datasets and learns accurate masks; 2) Transferability - the probabilistic modeling of mask distribution enables the transfer learning of sparsity across domains or tasks. The method achieves SOTA results on Wikitext and as well as shows lossless compression for many downstream language tasks. The code is available at https://github.com/NVlabs/MaskLLM.
- MRT5: DYNAMIC TOKEN MERGING FOR EFFICIENT BYTE-LEVEL LANGUAGE MODELS by Stanford University (https://arxiv.org/pdf/2410.20771). The paper introduces a more efficient variant of ByT5 that integrates a token deletion mechanism in its encoder to dynamically shorten the input sequence length. After processing through a fixed number of encoder layers, a learnt delete gate determines which tokens are to be removed and which are to be retained for subsequent layers. MrT5 effectively “merges” critical information from deleted tokens into a more compact sequence, leveraging contextual information from the remaining tokens. In continued pre-training experiments, we find that MrT5 can achieve significant gains in inference runtime with minimal effect on performance. Code is available here: https://github.com/jkallini/mrt5.
- SQFT: Low-cost Model Adaptation in Low-precision Sparse Foundation Models by Intel Labs (https://aclanthology.org/2024.findings-emnlp.749.pdf). The authors propose and end-to-end solution for low-precision sparse parameter-efficient fine-tuning of large pre-trained models, allowing for effective model adaptation in resource-constrained environments. Additionally, an innovative strategy enables the merging of sparse weights with low-rank adapters without losing sparsity and accuracy, overcoming the limitations of previous approaches. SQFT also addresses the challenge of having quantized weights and adapters with different numerical precisions, enabling merging in the desired numerical format without sacrificing accuracy. Multiple adaptation scenarios, models, and comprehensive sparsity levels demonstrate the effectiveness of SQFT. Models and code are available at https://github.com/IntelLabs/Hardware-Aware-Automated-Machine-Learning.
- Post-Training Statistical Calibration for Higher Activation Sparsity by Intel Labs (https://arxiv.org/pdf/2412.07174). The paper presents a post-training activation pruning framework that (1) generalizes sparsification by input activations of Fully-Connected layers for generic and flexible application across Transformers, and (2) features a simple Mode-Centering technique to pre-calibrate activation distributions for maximizing post-training sparsity. The results demonstrate robust Pareto efficiency compared to prior methods, translating to a 1.5x additional LLM decoding speedup against] at iso model quality. The effectiveness of the method is empirically verified across a wide range of models, including recent Transformer Decoders, MoE, Mamba2, Encoding Transformer, and pre-quantized models. The code is available at: https://github.com/IntelLabs/SCAP.
- HashAttention: Semantic Sparsity for Faster Inference by UC Berkeley and ETH Zurich (https://arxiv.org/pdf/2412.14468). The paper proposes an approach that is casting pivotal token identification as a recommendation problem. Given a query, it encodes keys and queries in Hamming space capturing the required semantic similarity using learned mapping functions. The method identifies pivotal tokens for a given query in this Hamming space using bitwise operations, and only these pivotal tokens are used for attention computation. It can reduce the number of tokens used by a factor of 1/32× for the Llama-3.1-8B model with LongBench, keeping average quality loss within 0.6 points, while using only 32 bits per token auxiliary memory. Code is planned to be released.
- BEYOND 2:4: EXPLORING V:N:M SPARSITY FOR EFFICIENT TRANSFORMER INFERENCE ON GPUS by Tsinghua University, Huawei Noah’s Ark Lab, Beijing Jiaotong University (https://arxiv.org/pdf/2410.16135). Authors propose three approaches to enhance the applicability and accuracy of V:N:M-sparse Transformers, including heuristic V and M selection, V:N:M-specific channel permutation and three-staged LoRA training techniques. Experimental results show that, with with this, the DeiT-small achieves lossless accuracy at 64:2:5 sparsity, while the DeiT-base maintains accuracy even at 64:2:8 sparsity. In addition, the fine-tuned LLama2-7B at 64:2:5 sparsity performs comparably or better than training-free 2:4 sparse alternatives on downstream tasks.
Other
- InfiniPot: Infinite Context Processing on Memory-Constrained LLMs from by Qualcomm AI Research , Qualcomm Korea YH (https://arxiv.org/pdf/2410.01518). The paper introduces a KV cache control framework designed to enable pre-trained LLMs to manage extensive sequences within fixed memory constraints efficiently, without requiring additional training. The method leverages Continual Context Distillation (CCD), an iterative process that compresses and retains essential information through novel importance metrics, maintaining critical data. This distillation process is based on the combination of CE-loss over the predicted tokens and Attention scores. Evaluations indicate that the method significantly outperforms models trained for long contexts in various NLP tasks.
- DEEP COMPRESSION AUTOENCODER FOR EFFICIENT HIGH-RESOLUTION DIFFUSION MODELS by MIT, Tsinghua University, and NVIDIA (https://arxiv.org/pdf/2410.10733). Authors highlight that existing autoencoders have demonstrated impressive results at a moderate spatial compression ratio (e.g., 8×) but fail to maintain satisfactory reconstruction accuracy for high spatial compression ratios (e.g., 64×). They address this by introducing two techniques: (1) Residual Autoencoding, where we design our models to learn residuals based on the space-to-channel transformed; (2) Decoupled High-Resolution Adaptation, a decoupled three-phase training strategy for mitigating the generalization penalty of high spatial-compression autoencoders. Authors improve the autoencoder’s spatial compression ratio up to 128 while maintaining the reconstruction quality achieving significant speedup without accuracy drop (19.1× inference speedup and 17.9× training speedup on H100 GPU). Code is available at https://github.com/mit-han-lab/efficientvit.
- EoRA: Training-free Compensation for Compressed LLM with Eigenspace Low-Rank Approximation by Nvidia (https://arxiv.org/pdf/2410.21271). The paper proposes a method that directly minimizes compression-induced errors without requiring gradient-based training small amount of calibration data. The method projects compression errors into the eigenspace of input activations, leveraging eigenvalues to effectively prioritize the reconstruction of high-importance error components. It shows good results for compressed LLaMA2/3 models on various tasks, such as language generation, commonsense reasoning, and math reasoning tasks (e.g., 31.31%/12.88% and 9.69% improvements on ARC-Easy/ARC-Challenge and MathQA when compensating LLaMA3-8B that is quantized to 4-bit and pruned to 2:4 sparsity).
- Eigen Attention: Attention in Low-Rank Space for KV Cache Compression by Purdue University (https://arxiv.org/pdf/2408.05646). Authors propose Eigen Attention, which performs the attention operation in a low-rank space, thereby reducing the KV cache memory overhead. The proposed approach is orthogonal to existing KV cache compression techniques and can be used synergistically with them. Experiments demonstrate that Eigen Attention results in up to 40% reduction in KV cache sizes and up to 60% reduction in attention operation latency with minimal drop in performance. Code is available at https://github.com/UtkarshSaxena1/EigenAttn.
- RAGCache: Efficient Knowledge Caching for Retrieval-Augmented Generation by Peking University and ByteDance (https://arxiv.org/pdf/2404.12457). Authors propose RAGCache, the system that caches the intermediate states of external knowledge and shares them across multiple queries to reduce the redundant computation. They design a prefix-aware GDSF replacement policy that leverages the characteristics of RAG to minimize the miss rate and a dynamic speculative pipelining approach to minimize the end-to-end latency. The experimental results show that RAGCache reduces the time to first token (TTFT) by up to 4× and improves the throughput by up to 2.1× compared to vLLM integrated with Faiss.
- STAR: Synthesis of Tailored Architectures by Liquid AI (https://arxiv.org/pdf/2411.17800). In this work, authors propose a NAS-based approach for the synthesis of LLM architectures. This approach combines a search space based on the theory of linear input-varying systems, supporting a hierarchical numerical encoding into architecture genomes. The genomes are automatically refined and recombined with gradient-free, evolutionary algorithms to optimize for multiple model quality and efficiency metrics. Using the method, authos optimize large populations of new architectures, leveraging diverse computational units and interconnection patterns, improving over highly-optimized Transformers and striped hybrid models on the frontier of quality, parameter size, and inference cache for autoregressive language modeling.
- SWITTI: Designing Scale-Wise Transformers for Text-to-Image Synthesis by Yandex Research, HSE University, MIPT, Skoltech, ITMO University (https://arxiv.org/pdf/2412.01819). The paper presents text-to-image transformer that employs architectural modifications to improve training stability and convergence and excludes explicit autoregression for more efficient sampling and better scalability. Compared to state-of-the-art text-to-image diffusion models, the model is up to 7× faster while demonstrating competitive performance. Additionally, the model reduces memory consumption during inference, previously needed for storing key-value (KV) cache, enabling better scaling to higher resolution image generation. The model has weaker reliance on the text at high-resolution scales. This observation allows to disable classifier-free guidance at the last two steps, resulting in further ∼20% acceleration and better generation of fine-grained details, as confirmed by human evaluation.
- SWIFTKV: FAST PREFILL-OPTIMIZED INFERENCE WITH KNOWLEDGE-PRESERVING MODEL TRANSFORMATION by Snowflake AI Research (https://arxiv.org/pdf/2410.03960). The paper presents a model transformation and distillation procedure specifically designed to reduce the time and cost of processing prompt tokens while preserving the quality of generated tokens. The method combines three key mechanisms: i) SingleInputKV, which prefills later layers’ KV cache using a much earlier layer’s output, allowing prompt tokens to skip much of the model computation, ii) AcrossKV, which merges the KV caches of neighboring layers to reduce the memory footprint and support larger batch size for higher throughput, and iii) a knowledge-preserving distillation to recover the accuracy. For Llama-3.1-8B and 70B, the method reduces the compute requirement of prefill by 50% and the memory requirement of the KV cache by 62.5% while incurring minimum quality degradation across a wide range of tasks. Optimized models are available here.
- KV PREDICTION FOR IMPROVED TIME TO FIRST TOKEN by Apple (https://arxiv.org/pdf/2410.08391). In this method, a small auxiliary model is used to process the prompt and produce an approximation of the KV cache used by a base model. This approximated KV cache is then used with the base model for autoregressive generation without the need to query the auxiliary model again. Authors demonstrate that the method produces a pareto-optimal efficiency-accuracy trade-off when compared to baselines. On TriviaQA, they demonstrate relative accuracy improvements in the range of 15%−50% across a range of TTFT FLOPs budgets. They also demonstrate accuracy improvements of up to 30% on HumanEval python code completion at fixed TTFT FLOPs budgets. We release our code here.
- MAMBAEXTEND: A TRAINING-FREE APPROACH TO IMPROVE LONG-CONTEXT EXTENSION OF MAMBA (https://openreview.net/pdf?id=LgzRo1RpLS). The paper discloses the method that aims to extend the context length of SSM models, in particular Mamba family. The method leverages a training-free approach to calibrate only the scaling factors of discretization modules for different layers. Authors demonstrate both gradient-based and gradient-free zeroth-order optimization to learn the optimal scaling factors for each Mamba layer, requiring orders of magnitude fewer updates as opposed to the parameter fine-tuning-based alternatives. The method shows good accuracy on the Pile and Longbench benchmarks.
- Exploiting LLM Quantization by ETH Zurich (https://arxiv.org/pdf/2405.18137). A method which produces a malicious LLM from an original LLM. Malicious model performs similarly while in FP32 precision but malicious after the quantization. LLM -> malicious LLM -> Repairing malicious LLM via projected gradient descent subject to quantization blocks of the malicious LLM
- DEEP COMPRESSION AUTOENCODER FOR EFFICIENT HIGH-RESOLUTION DIFFUSION MODELS by MIT, Tsinghua University, and NVIDIA (https://arxiv.org/pdf/2410.10733). The proposed method is aimed to optimize image generation autoencoders by introducing two key techniques: (1) Residual Autoencoding, where authors design models to learn residuals based on the space-to-channel transformed features to alleviate the optimization difficulty of high spatial-compression autoencoders; (2) Decoupled High-Resolution Adaptation, a decoupled three-phase training strategy for mitigating the generalization penalty of high spatial-compression autoencoders. The method improves the autoencoder’s spatial compression ratio up to 128 while maintaining the reconstruction quality. Authors achieve significant speedup without accuracy drop. For example, on ImageNet 512 × 512, the model provides 19.1× inference speedup and 17.9× training speedup on H100 GPU for UViT-H while achieving a better FID. Code is available at: https://github.com/mit-han-lab/efficientvit.
- DUOATTENTION: EFFICIENT LONG-CONTEXT LLM INFERENCE WITH RETRIEVAL AND STREAMING HEADS by MIT, Tsinghua University, SJTU, University of Edinburgh, NVIDIA (https://arxiv.org/pdf/2410.10819). In this paper, authors identify that only a fraction of attention heads, a.k.a, Retrieval Heads, are critical for processing long contexts and require full attention across all tokens. In contrast, all other heads, which primarily focus on recent tokens and attention sinks–referred to as Streaming Heads–do not require full attention. They introduce a framework that only applies a full KV cache to retrieval heads while using a light-weight, constant-length KV cache for streaming heads, which reduces both LLM’s decoding and pre-filling memory and latency. DuoAttention uses a lightweight, optimization-based algorithm with synthetic data to identify retrieval heads accurately. The method reduces long-context inference memory by up to 2.55× for MHA and 1.67× for GQA models while speeding up decoding by up to 2.18× and 1.50× and accelerating pre-filling by up to 1.73× and 1.63× for MHA and GQA models, respectively. Code is available at: https://github.com/mit-han-lab/duo-attention.
Software
- KV-COMPRESS: PAGED KV-CACHE COMPRESSION WITH VARIABLE COMPRESSION RATES PER ATTENTION HEAD by Cloudflare (https://arxiv.org/pdf/2410.00161). KV-Compress introduces a method to reduce the KV cache memory footprint by selectively compressing attention heads based on their importance. While early approaches measure KV importance by aggregating attention across all past queries, recent works show performance improvements by focusing on the final prompt tokens within a limited observation window. KV-Compress evicts contiguous KV blocks within a PagedAttention framework, reducing the memory footprint proportionally to the theoretical compression rate. Extending Ada-SnapKV, it supports per-layer and per-head variable compression rates, achieving state-of-the-art results on the LongBench suite. The "query-group-compression" technique further compresses the KV cache of GQA models without expanding it into the dimension of total query heads, achieving up to a 4x additional reduction. Integrated within vLLM, KV-Compress demonstrates the first end-to-end benchmarks of an eviction-based KV cache compression method within a paged-attention-enabled framework for efficient LLM inference. Code is available at https://github.com/IsaacRe/vllm-kvcompress.
- Introducing Machete, a Mixed-Input GEMM Kernel Optimized for NVIDIA Hopper GPUs.
- AMD released TensorCast, a casting/quantization PyTorch-based library to emulate various precisions: https://github.com/ROCm/tensorcast.
- MInference: Million-Tokens Prompt Inference for Long-context LLMs. A research project that is driven by Microsoft for a long-context text generation tasks. It contains implementation of several state-of-the-art methods.
Apply dynamic LoRA into Stable Diffusion v1.5 with OpenVINO
LoRA, or Low-Rank Adaptation, reduces the number of trainable parameters by learning pairs of rank-decompostion matrices while freezing the original weights. This vastly reduces the storage requirement for large language models adapted to specific tasks and enables efficient task-switching during deployment all without introducing inference latency. Thus for a basic large model, the task scenarios of the model can be changed by different LoRAs. In a previous blog, it has been described how to convert the LoRAs-fused base model from pytorch to OpenVINO IR, but this method has the shortcoming of not being able to dynamically switch between LoRAs, which happen to be famous for their flexibility.
This blog will introduce how to implement the dynamic switching of LoRAs in a trick way. Specifically, for most of the tasks, the structure of the base model and LoRAs is unchanged, what changes is the task-specific LoRAs weights, and we can use these LoRAs weights as inputs to the model to achieve the dynamic switching function. All the code involved in this blog can be found here.
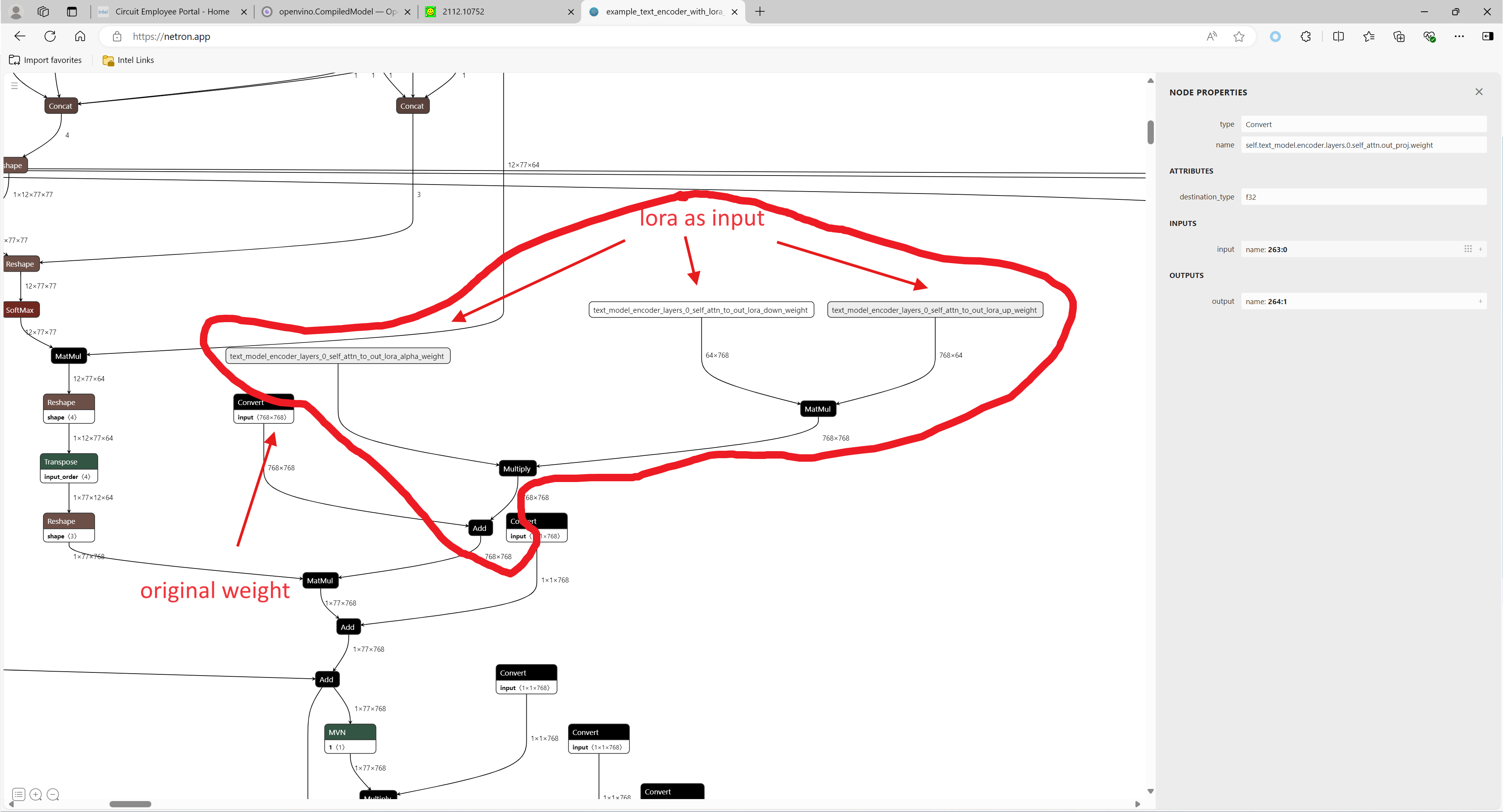
1. Environment preparation
# %python -m venv stable-diffusion-lora
# %source stable-diffusion-lora/bin/activate
git clone https://github.com/TianmengChen/sd1.5_controlnet_lora.git
pip install -r requirements.txt
2. Convert and inference
you should first change the lora file path and configs at first around line 478 in ov_model_export.py, after run python ov_model_ export.py, you will get related OpenVINO IR model. Then you can run ov_model_infer.py.
python ov_model_export.py
python ov_model_infer.py
3. Codes explanation
The most important part is the code in util.py, which is used to modify the model graph and load lora.
Function load_lora(lora_path, DEVICE_NAME) is used to load lora, get lora's shape and weights per layers and modify each layer's name.
def load_lora(lora_path, DEVICE_NAME):
state_dict = load_file(lora_path)
if DEVICE_NAME =="CPU":
for key, value in state_dict.items():
if isinstance(value, torch.Tensor):
value_fp32 = value.type(torch.float32)
state_dict[key] = value_fp32
layers_per_block = 2#TODO
state_dict = _maybe_map_sgm_blocks_to_diffusers(state_dict, layers_per_block)
state_dict, network_alphas = _convert_non_diffusers_lora_to_diffusers(state_dict)
# now keys in format like: "unet.up_blocks.0.attentions.2.transformer_blocks.8.ff.net.2.lora.down.weight"'
new_state_dict = {}
for key , value in state_dict.items():
if len(value.shape)==4:
# new_value = torch.reshape(value, (value.shape[0],value.shape[1]))
new_value = torch.squeeze(value)
else:
new_value = value
new_state_dict[key.replace('.', '_').replace('_processor','')] = new_value
# now keys in format like: "unet_up_blocks_0_attentions_2_transformer_blocks_8_ff_net_2_lora_down_weight"'
LORA_PREFIX_UNET = "unet"
LORA_PREFIX_TEXT_ENCODER = "text_encoder"
LORA_PREFIX_TEXT_2_ENCODER = "text_encoder_2"
lora_text_encoder_input_value_dict = {}
lora_text_encoder_2_input_value_dict = {}
lora_unet_input_value_dict = {}
lora_alpha = collections.Counter(network_alphas.values()).most_common()[0][0]
for key in new_state_dict.keys():
if LORA_PREFIX_TEXT_ENCODER in key and "lora_down" in key and LORA_PREFIX_TEXT_2_ENCODER not in key:
layer_infos = key.split(LORA_PREFIX_TEXT_ENCODER + "_")[-1]
lora_text_encoder_input_value_dict[layer_infos] = new_state_dict[key]
lora_text_encoder_input_value_dict[layer_infos.replace("lora_down", "lora_up")] = new_state_dict[key.replace("lora_down", "lora_up")]
elif LORA_PREFIX_TEXT_2_ENCODER in key and "lora_down" in key:
layer_infos = key.split(LORA_PREFIX_TEXT_2_ENCODER + "_")[-1]
lora_text_encoder_2_input_value_dict[layer_infos] = new_state_dict[key]
lora_text_encoder_2_input_value_dict[layer_infos.replace("lora_down", "lora_up")] = new_state_dict[key.replace("lora_down", "lora_up")]
elif LORA_PREFIX_UNET in key and "lora_down" in key:
layer_infos = key.split(LORA_PREFIX_UNET + "_")[-1]
lora_unet_input_value_dict[layer_infos] = new_state_dict[key]
lora_unet_input_value_dict[layer_infos.replace("lora_down", "lora_up")] = new_state_dict[key.replace("lora_down", "lora_up")]
#now the keys in format without prefix
return lora_text_encoder_input_value_dict, lora_text_encoder_2_input_value_dict, lora_unet_input_value_dict, lora_alpha
Function add_param(model, lora_input_value_dict) is used to add input parameter per names of related layers, which will be connected to model with manager.register_pass(InsertLoRAUnet(input_param_dict)) and manager.register_pass(InsertLoRATE(input_param_dict)), in these two classes, we search the whole model graph to find the related layers by their names and connect them with lora.
def add_param(model, lora_input_value_dict):
param_list = []
for key, value in lora_input_value_dict.items():
if '_lora_down' in key:
key_down = key
key_up = key_down.replace('_lora_down','_lora_up')
name_alpha = key_down.replace('_lora_down','_lora_alpha')
lora_alpha = ops.parameter(shape='',name=name_alpha)
lora_alpha.output(0).set_names({name_alpha})
# lora_down = ops.parameter(shape=[-1, lora_input_value_dict[key_down].shape[-1]], name=key_down)
lora_down = ops.parameter(shape=lora_input_value_dict[key_down].shape, name=key_down)
lora_down.output(0).set_names({key_down})
# lora_up = ops.parameter(shape=[lora_input_value_dict[key_up].shape[0], -1], name=key_up)
lora_up = ops.parameter(shape=lora_input_value_dict[key_up].shape, name=key_up)
lora_up.output(0).set_names({key_up})
param_list.append(lora_alpha)
param_list.append(lora_down)
param_list.append(lora_up)
model.add_parameters(param_list)
class InsertLoRAUnet(MatcherPass):
def __init__(self, input_param_dict):
MatcherPass.__init__(self)
self.model_changed = False
param = WrapType("opset10.Convert")
def callback(matcher: Matcher) -> bool:
root = matcher.get_match_root()
root_output = matcher.get_match_value()
for key in input_param_dict.keys():
if root.get_friendly_name().replace('.','_').replace('self_unet_','') == key.replace('_lora_down','').replace('to_out','to_out_0'):
key_down = key
key_up = key_down.replace('_lora_down','_lora_up')
key_alpha = key_down.replace('_lora_down','_lora_alpha')
consumers = root_output.get_target_inputs()
lora_up_node = input_param_dict.pop(key_up)
lora_down_node = input_param_dict.pop(key_down)
lora_alpha_node = input_param_dict.pop(key_alpha)
lora_weights = ops.matmul(data_a=lora_up_node, data_b=lora_down_node, transpose_a=False, transpose_b=False, name=key.replace('_down',''))
lora_weights_alpha = ops.multiply(lora_alpha_node, lora_weights)
if len(root.shape)!=len(lora_weights_alpha.shape):
# lora_weights_alpha_reshape = ops.reshape(lora_weights_alpha, root.shape, special_zero=False)
lora_weights_alpha_reshape = ops.unsqueeze(lora_weights_alpha, axes=[2, 3])
add_lora = ops.add(root,lora_weights_alpha_reshape,auto_broadcast='numpy')
else:
add_lora = ops.add(root,lora_weights_alpha,auto_broadcast='numpy')
for consumer in consumers:
consumer.replace_source_output(add_lora.output(0))
return True
# Root node wasn't replaced or changed
return False
self.register_matcher(Matcher(param,"InsertLoRAUnet"), callback)
class InsertLoRATE(MatcherPass):
def __init__(self, input_param_dict):
MatcherPass.__init__(self)
self.model_changed = False
param = WrapType("opset10.Convert")
def callback(matcher: Matcher) -> bool:
root = matcher.get_match_root()
root_output = matcher.get_match_value()
root_name = None
if 'Constant_' in root.get_friendly_name() and root.shape == ov.Shape([768,768]):
target_input = root.output(0).get_target_inputs()
for v in target_input:
for input_of_MatMul in v.get_node().inputs():
if input_of_MatMul.get_shape()== ov.Shape([1,77,768]):
Add_Node = input_of_MatMul.get_source_output().get_node()
for Add_Node_output in Add_Node.output(0).get_target_inputs():
if 'k_proj' in Add_Node_output.get_node().get_friendly_name():
for i in Add_Node_output.get_node().inputs():
if i.get_shape() == ov.Shape([768,768]) and 'k_proj' in i.get_source_output().get_node().get_friendly_name():
root_name = i.get_source_output().get_node().get_friendly_name().replace('k_proj', 'q_proj')
root_friendly_name = root_name if root_name else root.get_friendly_name()
for key in input_param_dict.keys():
if root_friendly_name.replace('.','_').replace('self_','') == key.replace('_lora_down','_proj').replace('_to','').replace('_self',''):
# print(root_friendly_name)
key_down = key
key_up = key_down.replace('_lora_down','_lora_up')
key_alpha = key_down.replace('_lora_down','_lora_alpha')
consumers = root_output.get_target_inputs()
lora_up_node = input_param_dict.pop(key_up)
lora_down_node = input_param_dict.pop(key_down)
lora_alpha_node = input_param_dict.pop(key_alpha)
lora_weights = ops.matmul(data_a=lora_up_node, data_b=lora_down_node, transpose_a=False, transpose_b=False, name=key.replace('_down',''))
lora_weights_alpha = ops.multiply(lora_alpha_node, lora_weights)
add_lora = ops.add(root,lora_weights_alpha,auto_broadcast='numpy')
for consumer in consumers:
consumer.replace_source_output(add_lora.output(0))
return True
if len(input_param_dict) == 0:
print("All loras are added")
# Root node wasn't replaced or changed
return False
self.register_matcher(Matcher(param,"InsertLoRATE"), callback)
4. GenAI
In addition to this, the latest OpenVINO GenAI provides the Cpp API for LoRA. You can find it here.
OpenVINO GenAI Serving (OGS) update
Authors: Xiake Sun, Su Yang, Tianmeng Chen, Tong Qiu
OpenVINO GenAI Server (OGS) Update:
-Update LLM: stream generation, reset handle, multi-round chat, model cache config
-Support VLM
-Support Reranker for RAG sample
-Support BLIP image embedding for photo search with DB
-Support C++ GUI with imgui for photo search
Now we scale the text embedding to image embedding for RAG sample and support multi-Vector Retriever for RAG.
- Multi-Vector Retriever for RAG on text: QA over Document
- Multi-Vector Retriever for RAG on image: Photo search with DB retrieval
Here is a photo search sample with image embedding.
Usage 2: Photo Search with DB retrieval
Steps:
1.use python client to create image vector DB (PostgreSQL)
2.use GUI to search image
Here is a sample image to demonstrate GUI usage on client platform. we search the bus photo with top 10 similar images from the 100 images which are embedded into Vector DB.
Usage 3: Chat with images via MiniCPM-V
Once we have created a multimodal vector DB through image embedding, we can further communicate with the image through VLM.
We integrate the C++ GenAI sample visual_language_chat with openbmb/MiniCPM-V-2_6.
Here is the demo image on client platform.
OpenVINO GenAI Serving (OGS)
Authors: Fiona Zhao, Xiake Sun, Wenyi Zou, Su Yang, Tianmeng Chen
Model Server reference implementation based on OpenVINO GenAI Package for Edge/Client AI PC Use Case.
Use Case 1: C++ RAG Sample that supports most popular models like LLaMA 2
This example showcases for Retrieval-Augmented Generation based on text-generation Large Language Models (LLMs): chatglm, LLaMA, Qwen and other models with the same signature and Bert model for embedding feature extraction. The sample fearures ov::genai::LLMPipeline and configures it for the chat scenario. There is also a Jupyter notebook which provides an example of LLM-powered RAG in Python.
Download and convert the model and tokenizers
The --upgrade-strategy eager option is needed to ensure optimum-intel is upgraded to the latest version.
Setup of PostgreSQL, Libpqxx and Pgvector
Langchain's document Loader and Spliter
- Load: document_loaders is used to load document data.
- Split: text_splitter breaks large Documents into smaller chunks. This is useful both for indexing data and for passing it in to a model, since large chunks are harder to search over and won’t in a model’s finite context window.
PostgreSQL
Download postgresql from enterprisedb.(postgresql-16.2-1-windows-x64.exe is tested)
Install PostgreSQL with postgresqltutorial.
Setup of PostgreSQL:
1. Open pgAdmin 4 from Windows Search Bar.
2. Click Browser (left side) > Servers > Postgre SQL 10.
3. Create the user postgres with password openvino (or your own setting)
4. Open SQL Shell from Windows Search Bar to check this setup. 'Enter' to set Server, Database, Port, Username as default and type Password.
libpqxx
'Official' C++ client library (language binding), built on top of C library
Update the source code from https://github.com/jtv/libpqxx in deps\libpqxx
The pipeline connects with DB based on Libpqxx.
pgvector
Open-source vector similarity search for Postgres.
By default, pgvector performs exact nearest neighbor search, which provides perfect recall. It also supports approximate nearest neighbor search (HNSW), which trades some recall for speed.
For Windows, Ensure C++ support in Visual Studio 2022 is installed, then use nmake to build in Command Prompt for VS 2022(run as Administrator). Please follow with the pgvector
Enable the extension (do this once in each database where you want to use it), run SQL Shell from Windows Search Bar with "CREATE EXTENSION vector;".
Printing CREATE EXTENSION shows successful setup of Pgvector.
pgvector-cpp
pgvector support for C++ (supports libpqxx). The headers (pqxx.hpp, vector.hpp, halfvec.hpp) are copied into the local folder rag_sample\include. Our pipeline does the vector similarity search for the chunks embeddings in PostgreSQL, based on pgvector-cpp.
Install OpenVINO, VS2022 and Build this pipeline
Download 2024.2 release from OpenVINO™ archives*. This OV built package is for C++ OpenVINO pipeline, no need to build the source code. Install latest Visual Studio 2022 Community for the C++ dependencies and LLM C++ pipeline editing.
Extract the zip file in any location and set the environment variables with dragging this setupvars.bat in the terminal Command Prompt. setupvars.ps1 is used for terminal PowerShell. <INSTALL_DIR> below refers to the extraction location. Run the following CMD in the terminal Command Prompt.
Notice:
- Install on Windows: Copy all the DLL files of PostgreSQL, OpenVINO and tbb and openvino-genai into the release folder. The SQL DLL files locate in the installed PostgreSQL path like "C:\Program Files\PostgreSQL\16\bin".
- If cmake not installed in the terminal Command Prompt, please use the terminal Developer Command Prompt for VS 2022 instead.
- The openvino tokenizer in the third party needs several minutes to build. Set 8 for -j option to specify the number of parallel jobs.
- Once the cmake finishes, check rag_sample_client.exe and rag_sample_server.exe in the relative path .\build\samples\cpp\rag_sample\Release.
- If Cmake completed without errors, but not find exe, please open the .\build\OpenVINOGenAI.sln in VS2022, and set the solution configuration as Release instead of Debug, then build the llm project within VS2022 again.
Run
Launch RAG Server
rag_sample_server.exe --llm_model_path TinyLlama-1.1B-Chat-v1.0 --llm_device CPU --embedding_model_path bge-large-zh-v1.5 --embedding_device CPU --db_connection "user=postgres host=localhost password=openvino port=5432 dbname=postgres"
Lanuch RAG Client
rag_sample_client.exe
Lanuch python Client
Use python client to send the message of DB init and send the document chunks to DB for embedding and storing.
python client_get_chunks_embeddings.py --docs test_document_README.md